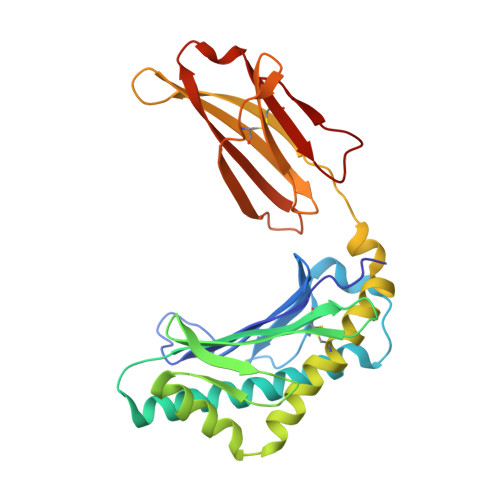
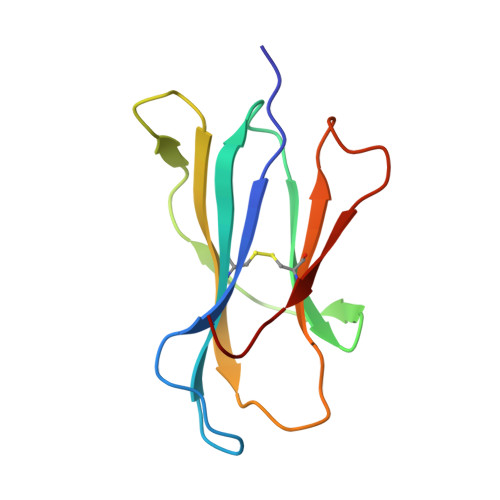
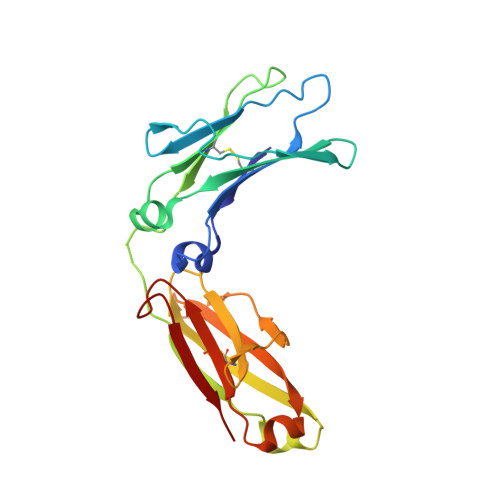
Crystal structure of the complex of rat neonatal Fc receptor with Fc.
Burmeister, W.P., Huber, A.H., Bjorkman, P.J.(1994) Nature 372: 379-383
- PubMed: 7969498
- DOI: https://doi.org/10.1038/372379a0
- Primary Citation of Related Structures:
1FRT - PubMed Abstract:
The neonatal Fc receptor (FcRn) transports maternal immunoglobulin G (IgG) to the bloodstream of the newborn. FcRn is structurally similar to class I major histocompatibility complex (MHC) molecules, despite differences in the ligands they bind (the Fc portion of IgG and antigenic peptides, respectively). A low-resolution crystal structure of the complex between FcRn and Fc localizes the binding site for Fc to the side of FcRn, distinct from the tops of the alpha 1 and alpha 2 domains which serve as the peptide and T-cell receptor binding sites in class I molecules. FcRn binds to Fc at the interface between the Fc CH2 and CH3 domains, which contains several histidine residues that could account for the sharply pH-dependent FcRn/IgG interaction. A dimer of FcRn heterodimers observed in the co-crystals and in the crystals of FcRn alone could be involved in binding Fc, correlating with the 2:1 binding stoichiometry between FcRn and IgG (ref. 4) and suggesting an unusual orientation of FcRn on the membrane.
- Division of Biology 156-29, California Institute of Technology, Pasadena 91125.
Organizational Affiliation: