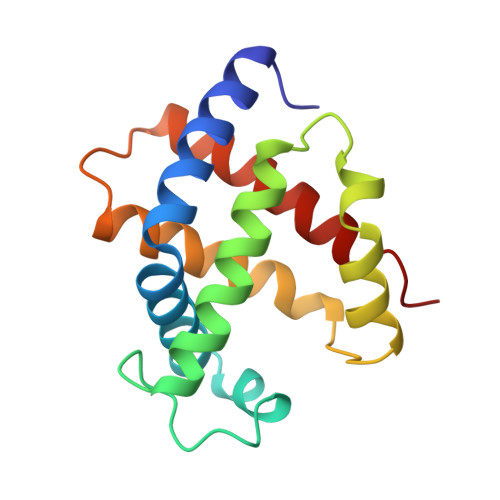
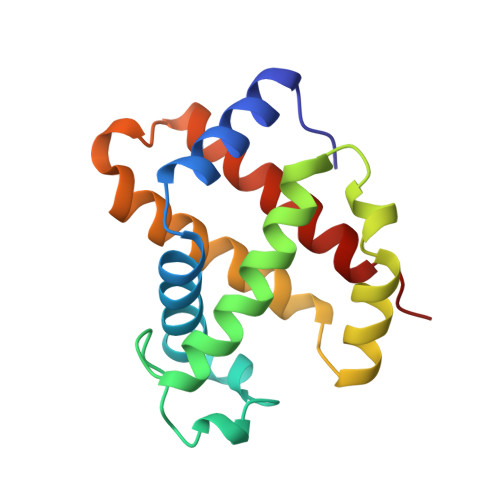
Structures of the deoxy and CO forms of haemoglobin from Dasyatis akajei, a cartilaginous fish.
Chong, K.T., Miyazaki, G., Morimoto, H., Oda, Y., Park, S.Y.(1999) Acta Crystallogr D Biol Crystallogr 55: 1291-1300
- PubMed: 10393295
- DOI: https://doi.org/10.1107/s0907444999005934
- Primary Citation of Related Structures:
1CG5, 1CG8 - PubMed Abstract:
The three-dimensional structures of the deoxy- and carbonmonoxyhaemoglobin (Hb) from Dasyatis akajei, a stingray, have been determined at 1.6 and 1.9 A resolution, respectively. This is one of the most distantly related vertebrate Hbs to human HbA. Both structures resemble the respective forms of HbA, indicating that the alpha2beta2-type tetramer and the mode of the quaternary structure change are common to Hbs of jawed vertebrates. Larger deviations between D. akajei Hb and human HbA are observed in various parts of the molecule, even in the E and F helices. Significant mutations and/or conformational changes are also observed around the haems, in the C-terminal region of the beta subunit, in the alpha1beta2 interface and in the organic phosphate-binding site of HbA. Despite these structural differences, the oxygen affinity, haem-haem interaction, Bohr effect and organic phosphate effect of D. akajei Hb are all only moderately reduced. Compared with human HbA, the overall r.m.s. deviation of main-chain atoms in the helical regions of bony fish Hbs is smaller than that of D. akajei Hb.
- Division of Biophysical Engineering, Graduate School of Engineering Science, Osaka University, Toyonaka, Osaka 560-8531, Japan.
Organizational Affiliation: