Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth.
Huang, C.C., Stricher, F., Martin, L., Decker, J.M., Majeed, S., Barthe, P., Hendrickson, W.A., Robinson, J., Roumestand, C., Sodroski, J., Wyatt, R., Shaw, G.M., Vita, C., Kwong, P.D.(2005) Structure 13: 755-768
- PubMed: 15893666
- DOI: https://doi.org/10.1016/j.str.2005.03.006
- Primary Citation of Related Structures:
1YYL, 1YYM - PubMed Abstract:
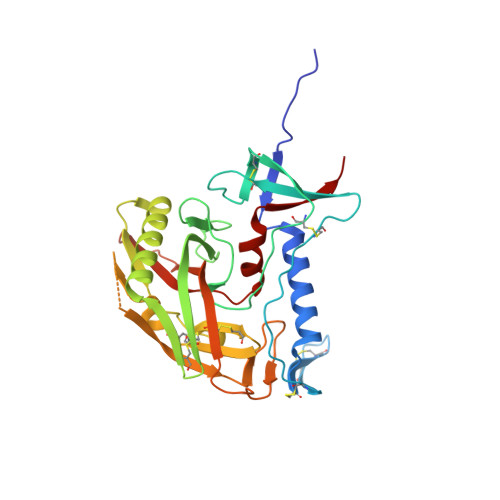
The binding surface on CD4 for the HIV-1 gp120 envelope glycoprotein has been transplanted previously onto a scorpion-toxin scaffold. Here, we use X-ray crystallography to characterize atomic-level details of gp120 with this transplant, CD4M33. Despite known envelope flexibility, the conformation of gp120 induced by CD4M33 was so similar to that induced by CD4 that localized measures were required to distinguish ligand-induced differences from lattice variation. To investigate relationships between structure, function, and mimicry, an F23 analog of CD4M33 was devised. Structural and thermodynamic analyses showed F23 to be a better molecular mimic of CD4 than CD4M33. F23 also showed increased neutralization breadth, against diverse isolates of HIV-1, HIV-2, and SIVcpz. Our results lend insight into the stability of the CD4 bound conformation of gp120, define measures that quantify molecular mimicry as a function of evolutionary distance, and suggest how such evaluations might be useful in developing mimetic antagonists with increased neutralization breadth.
- Vaccine Research Center, National Institutes of Health, Bethesda, Maryland 20892, USA.
Organizational Affiliation: