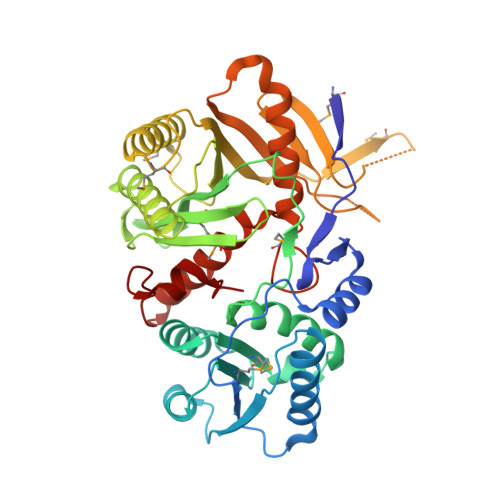
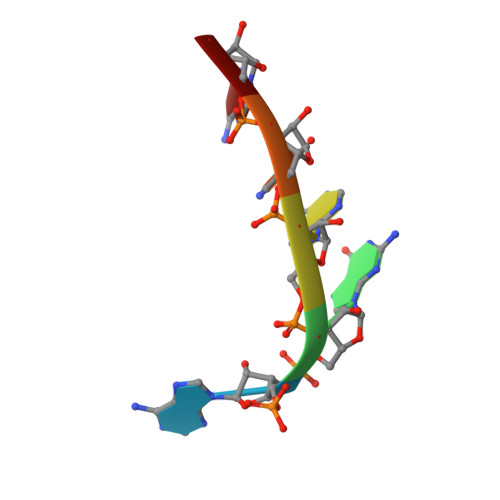
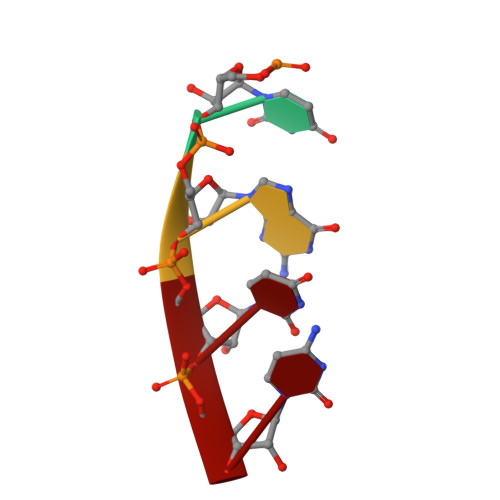
Structural basis for 5'-end-specific recognition of guide RNA by the A. fulgidus Piwi protein.
Ma, J.B., Yuan, Y.R., Meister, G., Pei, Y., Tuschl, T., Patel, D.J.(2005) Nature 434: 666-670
- PubMed: 15800629
- DOI: https://doi.org/10.1038/nature03514
- Primary Citation of Related Structures:
1YTU - PubMed Abstract:
RNA interference (RNAi) is a conserved sequence-specific gene regulatory mechanism mediated by the RNA-induced silencing complex (RISC), which is composed of a single-stranded guide RNA and an Argonaute protein. The PIWI domain, a highly conserved motif within Argonaute, has been shown to adopt an RNase H fold critical for the endonuclease cleavage activity of RISC. Here we report the crystal structure of Archaeoglobus fulgidus Piwi protein bound to double-stranded RNA, thereby identifying the binding pocket for guide-strand 5'-end recognition and providing insight into guide-strand-mediated messenger RNA target recognition. The phosphorylated 5' end of the guide RNA is anchored within a highly conserved basic pocket, supplemented by the carboxy-terminal carboxylate and a bound divalent cation. The first nucleotide from the 5' end of the guide RNA is unpaired and stacks over a conserved tyrosine residue, whereas successive nucleotides form a four-base-pair RNA duplex. Mutation of the corresponding amino acids that contact the 5' phosphate in human Ago2 resulted in attenuated mRNA cleavage activity. Our structure of the Piwi-RNA complex, and that determined elsewhere, provide direct support for the 5' region of the guide RNA serving as a nucleation site for pairing with target mRNA and for a fixed distance separating the RISC-mediated mRNA cleavage site from the anchored 5' end of the guide RNA.
- Structural Biology Program, Memorial Sloan-Kettering Cancer Center, New York, New York 10021, USA.
Organizational Affiliation: