Design and X-ray crystal structures of human thrombin with synthetic cyanopeptide-analogues.
Radau, G., Fokkens, J.(2007) Pharmazie 62: 83-88
- PubMed: 17341023
- Primary Citation of Related Structures:
1YPL, 1YPM - PubMed Abstract:
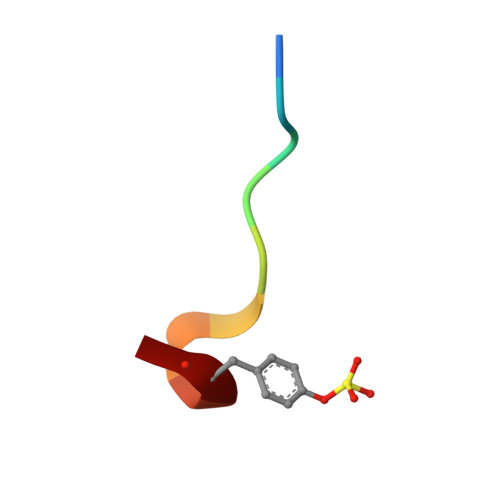
Based on the X-ray crystals of cocrystallized cyanopeptide-trypsin and cyanopeptide-thrombin-com-plexes, a rational drug design succeeded in the establishment of suitable lead structures for the development of new potential inhibitors of thrombin. This report deals with the design and X-ray crystallography data of new synthetic, low-molecular weight cyanopeptide-analogues, RA-1008 and RA-1014, complexed with human alpha-thrombin at 1.85 A resolution. The crystal structures of the complexes reveal, by analogy with modeling studies, that the salt bridge of Asp189 to this type of synthetic thrombin inhibitors leads to an almost identically binding into the S1 specificity pocket in comparison to the complex of the natural products, whereas in the overall binding modes the P2-P4 substructures differ from those of the leads. The strongest member of the second series of described thrombin inhibitors, RA-1014, shows in the crystal complex with thrombin a slightly higher affinity towards the enzyme than RA-1008 as confirmed by inhibition tests. This result and other key informations will be helpful to design a more potent series of inhibitors.
- Institute of Pharmacy, Ernst-Moritz-Arndt-University Greifswald, Germany. radau@uni-greifswald.de
Organizational Affiliation: