X-ray crystallography on ribosome recycling: mechanism of binding and action of RRF on the 50S ribosomal subunit
Wilson, D.N., Schluenzen, F., Harms, J.M., Yoshida, T., Ohkubo, T., Albrecht, R., Buerger, J., Kobayashi, Y., Fucini, P.(2005) EMBO J 24: 251-260
- PubMed: 15616575
- DOI: https://doi.org/10.1038/sj.emboj.7600525
- Primary Citation of Related Structures:
1Y69 - PubMed Abstract:
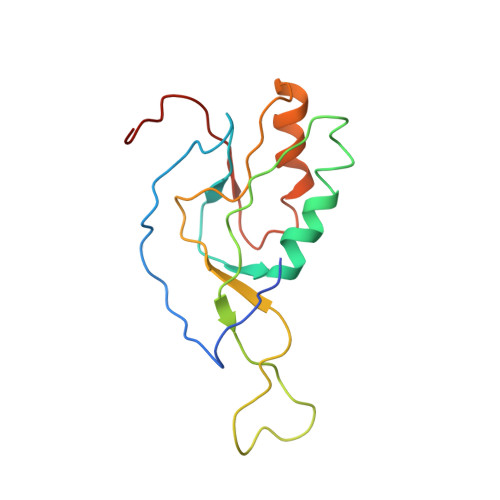
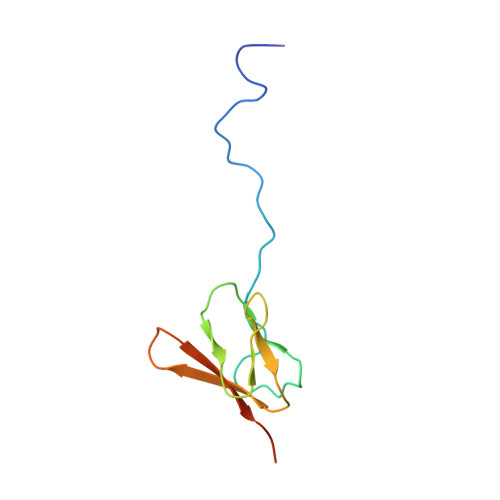
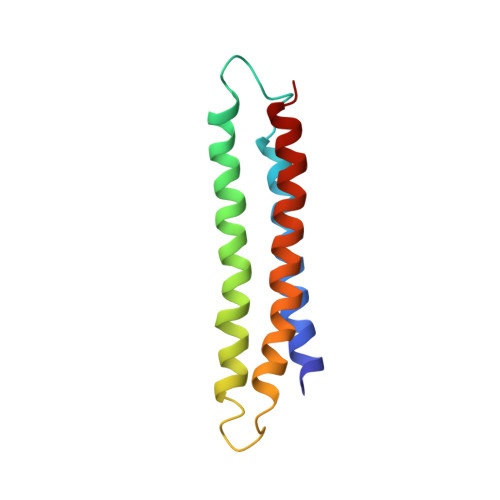
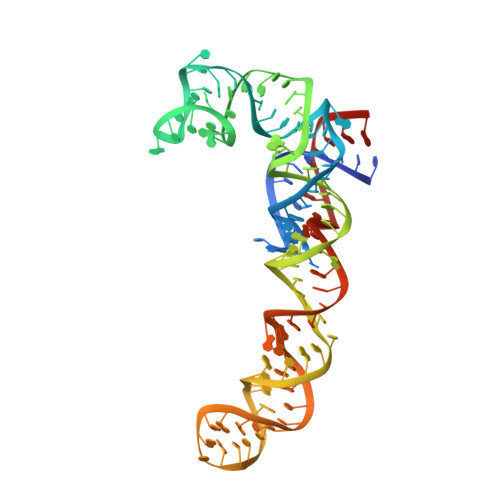
This study presents the crystal structure of domain I of the Escherichia coli ribosome recycling factor (RRF) bound to the Deinococcus radiodurans 50S subunit. The orientation of RRF is consistent with the position determined on a 70S-RRF complex by cryoelectron microscopy (cryo-EM). Alignment, however, requires a rotation of 7 degrees and a shift of the cryo-EM RRF by a complete turn of an alpha-helix, redefining the contacts established with ribosomal components. At 3.3 A resolution, RRF is seen to interact exclusively with ribosomal elements associated with tRNA binding and/or translocation. Furthermore, these results now provide a high-resolution structural description of the conformational changes that were suspected to occur on the 70S-RRF complex, which has implications for the synergistic action of RRF with elongation factor G (EF-G). Specifically, the tip of the universal bridge element H69 is shifted by 20 A toward h44 of the 30S subunit, suggesting that RRF primes the intersubunit bridge B2a for the action of EF-G. Collectively, our data enable a model to be proposed for the dual action of EF-G and RRF during ribosome recycling.
- Max-Planck-Institute for Molecular Genetics, Berlin, Germany.
Organizational Affiliation: