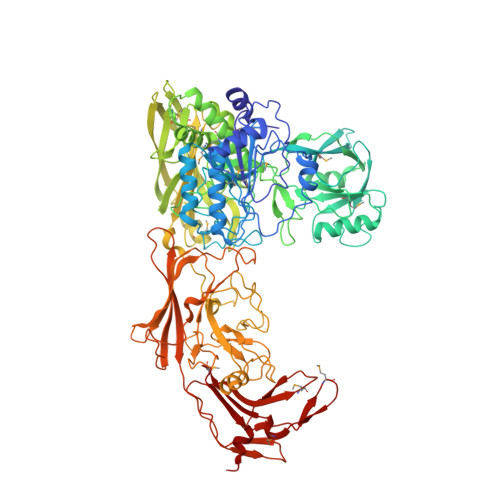
Structure of the streptococcal cell wall C5a peptidase
Brown, C.K., Gu, Z.Y., Matsuka, Y.V., Purushothaman, S.S., Winter, L.A., Cleary, P.P., Olmsted, S.B., Ohlendorf, D.H., Earhart, C.A.(2005) Proc Natl Acad Sci U S A 102: 18391-18396
- PubMed: 16344483
- DOI: https://doi.org/10.1073/pnas.0504954102
- Primary Citation of Related Structures:
1XF1 - PubMed Abstract:
The structure of a cell surface enzyme from a gram-positive pathogen has been determined to 2-A resolution. Gram-positive pathogens have a thick cell wall to which proteins and carbohydrate are covalently attached. Streptococcal C5a peptidase (SCP), is a highly specific protease and adhesin/invasin. Structural analysis of a 949-residue fragment of the [D130A,S512A] mutant of SCP from group B Streptococcus (S. agalactiae, SCPB) revealed SCPB is composed of five distinct domains. The N-terminal subtilisin-like protease domain has a 134-residue protease-associated domain inserted into a loop between two beta-strands. This domain also contains one of two Arg-Gly-Asp (RGD) sequences found in SCPB. At the C terminus are three fibronectin type III (Fn) domains. The second RGD sequence is located between Fn1 and Fn2. Our analysis suggests that SCP binding to integrins by the RGD motifs may stabilize conformational changes required for substrate binding.
- Department of Biochemistry, Molecular Biology, and Biophysics, University of Minnesota, Minneapolis, MN 55455, USA.
Organizational Affiliation: