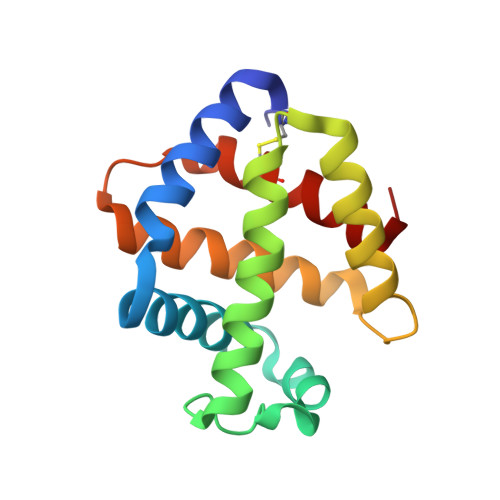
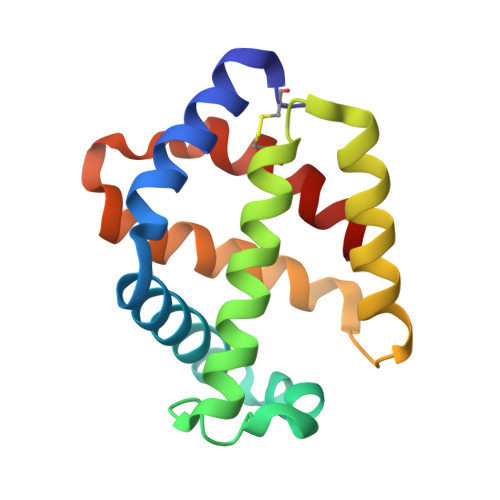
Crystal structure of the hemoglobin dodecamer from lumbricus erythrocruorin: allosteric core of giant annelid respiratory complexes
Strand, K., Knapp, J.E., Bhyravbhatla, B., Royer Jr., W.E.(2004) J Mol Biology 344: 119-134
- PubMed: 15504406
- DOI: https://doi.org/10.1016/j.jmb.2004.08.094
- Primary Citation of Related Structures:
1X9F - PubMed Abstract:
Erythrocruorins are highly cooperative giant extracellular respiratory complexes found in annelids, where they serve the same function as red blood cells. Our previous 5.5A resolution crystal structure of Lumbricus terrestris erythrocruorin revealed a hierarchical organization of 144 oxygen-binding hemoglobin chains that are assembled into 12 dodecamers arranged at the periphery of the complex around a central scaffold formed by 36 non-hemoglobin subunits. We present here the 2.6A resolution crystal structure of the Lumbricus hemoglobin dodecameric subassembly, which provides the first atomic models of the erythrocruorin allosteric core. The hemoglobin dodecamer has a molecular 3-fold axis of symmetry that relates three heterotetramers, each of which is composed of two tightly associated heterodimers. The structure reveals details of the interfaces, including key side-chain interactions likely to contribute to ligand-linked allosteric transitions, and shows the crowded nature of the ligand-binding pockets. Comparison of the Lumbricus dimeric assemblies with similar ones from mollusks and echinoderms suggests plausible pH-dependent quaternary transitions that may occur in response to proton binding and ligand release. Thus, these results provide the first step towards elucidating the structural basis for the strong allosteric properties of Lumbricus erythrocruorin.
- Department of Biochemistry and Molecular Pharmacology, University of Massachusetts Medical School, LRB 921, 364 Plantation Street, Worcester, MA 01605, USA.
Organizational Affiliation: