Interaction of Era with the 30S Ribosomal Subunit Implications for 30S Subunit Assembly
Sharma, M.R., Barat, C., Wilson, D.N., Booth, T.M., Kawazoe, M., Hori-Takemoto, C., Shirouzu, M., Yokoyama, S., Fucini, P., Agrawal, R.K.(2005) Mol Cell 18: 319-329
- PubMed: 15866174
- DOI: https://doi.org/10.1016/j.molcel.2005.03.028
- Primary Citation of Related Structures:
1X18, 1X1L - PubMed Abstract:
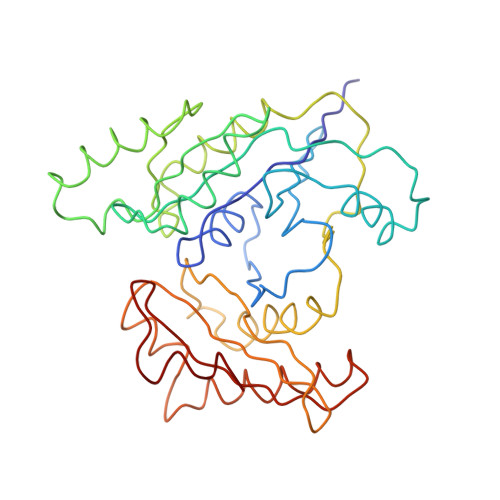
Era (E. coliRas-like protein) is a highly conserved and essential GTPase in bacteria. It binds to the 16S ribosomal RNA (rRNA) of the small (30S) ribosomal subunit, and its depletion leads to accumulation of an unprocessed precursor of the 16S rRNA. We have obtained a three-dimensional cryo-electron microscopic map of the Thermus thermophilus 30S-Era complex. Era binds in the cleft between the head and platform of the 30S subunit and locks the subunit in a conformation that is not favorable for association with the large (50S) ribosomal subunit. The RNA binding KH motif present within the C-terminal domain of Era interacts with the conserved nucleotides in the 3' region of the 16S rRNA. Furthermore, Era makes contact with several assembly elements of the 30S subunit. These observations suggest a direct involvement of Era in the assembly and maturation of the 30S subunit.
- Division of Molecular Medicine, Wadsworth Center, New York State Department of Health, Empire State Plaza, Albany, New York 12201, USA.
Organizational Affiliation: