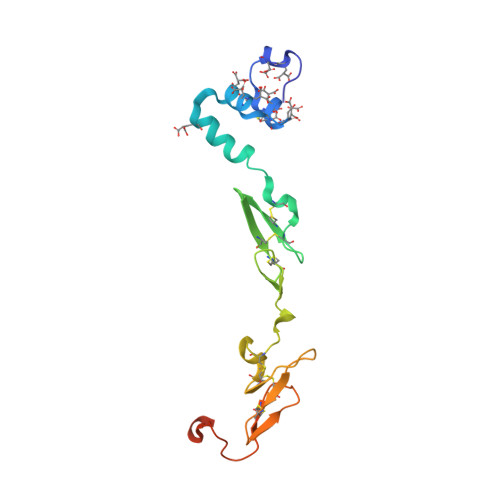
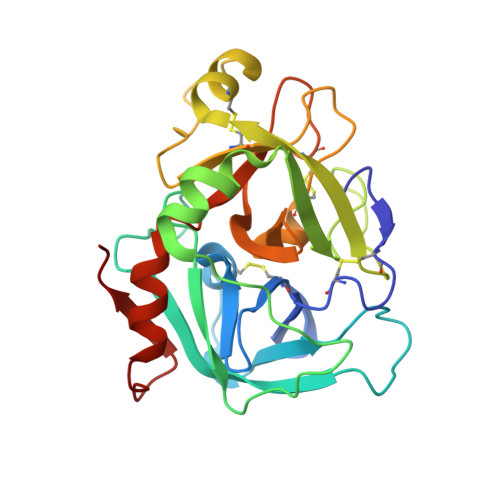
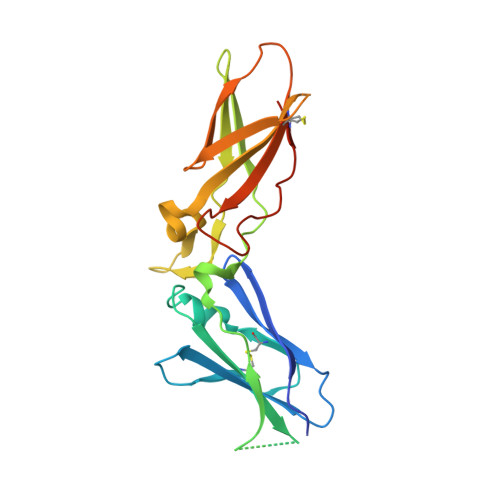
Crystal structure of human factor VIIa/tissue factor in complex with peptide mimetic inhibitor
Kadono, S., Sakamoto, A., Kikuchi, Y., Oh-eda, M., Yabuta, N., Koga, T., Hattori, K., Shiraishi, T., Haramura, M., Kodama, H., Esaki, T., Sato, H., Watanabe, Y., Itoh, S., Ohta, M., Kozono, T.(2004) Biochem Biophys Res Commun 324: 1227-1233
- PubMed: 15504346
- DOI: https://doi.org/10.1016/j.bbrc.2004.09.182
- Primary Citation of Related Structures:
1WQV - PubMed Abstract:
The 3D structure of human factor VIIa/soluble tissue factor in complex with a peptide mimetic inhibitor, propylsulfonamide-D-Thr-Met-p-aminobenzamidine, is determined by X-ray crystallography. As compared with the interactions between thrombin and thrombin inhibitors, the interactions at S2 and S3 sites characteristic of factor VIIa and factor VIIa inhibitors are revealed. The S2 site has a small pocket, which is filled by the hydrophobic methionine side chain in P2. The small S3 site fits the small size residue, D-threonine in P3. The structural data and SAR data of the peptide mimetic inhibitor show that these interactions in the S2 and S3 sites play an important role for the improvement of selectivity versus thrombin. The results will provide valuable information for the structure-based drug design of specific inhibitors for FVIIa/TF.
- Fuji Gotemba Research Labs, Chugai Pharmaceutical Co., Ltd., 1-135 Komakado, Gotemba, Shizuoka 412-8513, Japan. kadonosuj@chugai-pharm.co.jp
Organizational Affiliation: