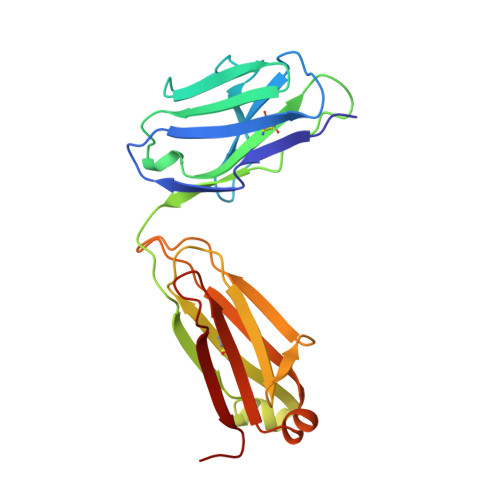
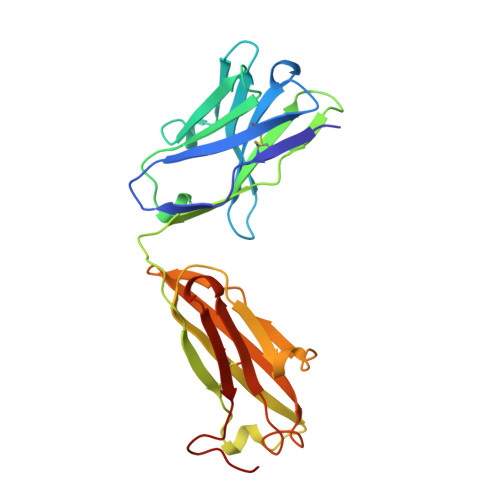
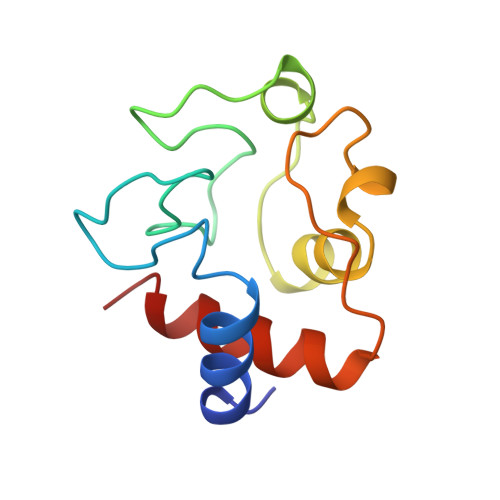
Structural basis for the binding of an anti-cytochrome c antibody to its antigen: crystal structures of FabE8-cytochrome c complex to 1.8 A resolution and FabE8 to 2.26 A resolution.
Mylvaganam, S.E., Paterson, Y., Getzoff, E.D.(1998) J Mol Biology 281: 301-322
- PubMed: 9698550
- DOI: https://doi.org/10.1006/jmbi.1998.1942
- Primary Citation of Related Structures:
1QBL, 1QBM, 1WEJ - PubMed Abstract:
A complete understanding of antibody-antigen association and specificity requires the stereochemical description of both antigen and antibody before and upon complex formation. The structural mechanism involved in the binding of the IgG1 monoclonal antibody E8 to its highly charged protein antigen horse cytochrome c (cyt c) is revealed by crystallographic structures of the antigen-binding fragment (Fab) of E8 bound to cyt c (FabE8-cytc), determined to 1.8 A resolution, and of uncomplexed Fab E8 (FabE8), determined to 2.26 A resolution. E8 antibody binds to three major discontiguous segments (33 to 39; 56 to 66; 96 to 104), and two minor sites on cyt c opposite to the exposed haem edge. Crystallographic definition of the E8 epitope complements and extends biochemical mapping and two-dimensional nuclear magnetic resonance with hydrogen-deuterium exchange studies. These combined results demonstrate that antibody-induced stabilization of secondary structural elements within the antigen can propagate locally to adjacent residues outside the epitope. Pre-existing shape complementarity at the FabE8-cytc interface is enhanced by 48 bound water molecules, and by local movements of up to 4.2 A for E8 antibody and 8.9 A for cyt c. Glu62, Asn103 and the C-terminal Glu104 of cyt c adjust to fit the pre-formed VL "hill" and VH "valley" shape of the grooved E8 paratope. All six E8 complementarity determining regions (CDRs) contact the antigen, with CDR L1 forming 46% of the total atomic contacts, and CDRs L1 (29%) and H3 (20%) contributing the highest percentage of the total surface area of E8 buried by cyt c (550 A2). The E8 antibody covers 534 A2 of the cyt c surface. The formation of five ion pairs between E8 and flexible cyt c residues Lys60, Glu62 and Glu104 suggests the importance of mobile regions and electrostatic interactions in providing the exquisite specificity needed for recognition of this extremely conserved protein antigen. The highly homologous VL domains of E8 and anti-lysozyme antibody D1. 3 achieve their distinct antigen-binding specificities by expanding the impact of their limited sequence differences through the recruitment of different sets of conserved residues and distinctly different CDR L3 conformations.
- Department of Molecular Biology and the Skaggs Institute for Chemical Biology, The Scripps Research Institute, 10550 N. Torrey Pines Rd, La Jolla, CA 92037, USA. shankari@scripps.edu
Organizational Affiliation: