Dual Role of Muts Glutamate 38 in DNA Mismatch Discrimination and in the Authorization of Repair.
Lebbink, J.H.G., Georgijevic, D., Natrajan, G., Fish, A., Winterwerp, H.H.K., Sixma, T.K., De Wind, N.(2006) EMBO J 25: 409
- PubMed: 16407973
- DOI: https://doi.org/10.1038/sj.emboj.7600936
- Primary Citation of Related Structures:
1WB9, 1WBB, 1WBD - PubMed Abstract:
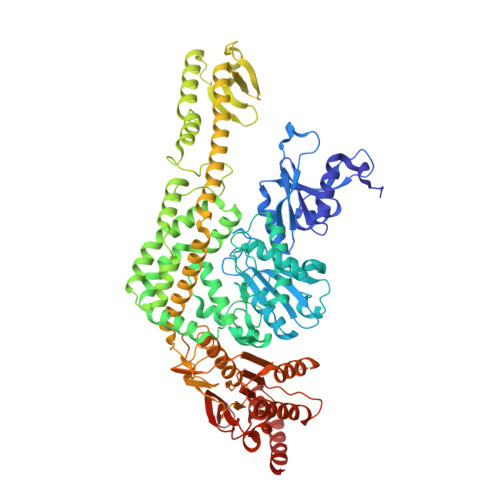
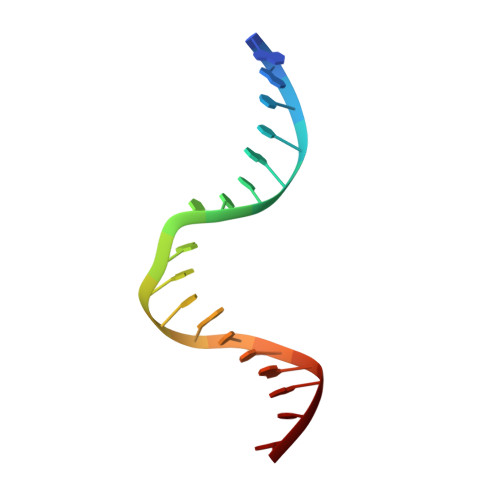
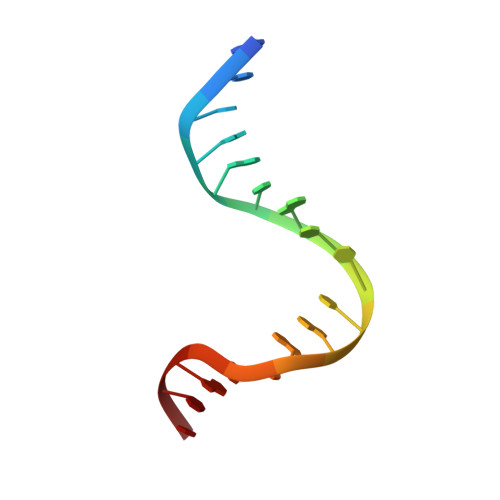
MutS plays a critical role in DNA mismatch repair in Escherichia coli by binding to mismatches and initiating repair in an ATP-dependent manner. Mutational analysis of a highly conserved glutamate, Glu38, has revealed its role in mismatch recognition by enabling MutS to discriminate between homoduplex and mismatched DNA. Crystal structures of MutS have shown that Glu38 forms a hydrogen bond to one of the mismatched bases. In this study, we have analyzed the crystal structures, DNA binding and the response to ATP binding of three Glu38 mutants. While confirming the role of the negative charge in initial discrimination, we show that in vivo mismatch repair can proceed even when discrimination is low. We demonstrate that the formation of a hydrogen bond by residue 38 to the mismatched base authorizes repair by inducing intramolecular signaling, which results in the inhibition of rapid hydrolysis of distally bound ATP. This allows formation of the stable MutS-ATP-DNA clamp, a key intermediate in triggering downstream repair events.
- Division of Molecular Carcinogenesis, The Netherlands Cancer Institute, Amsterdam, The Netherlands.
Organizational Affiliation: