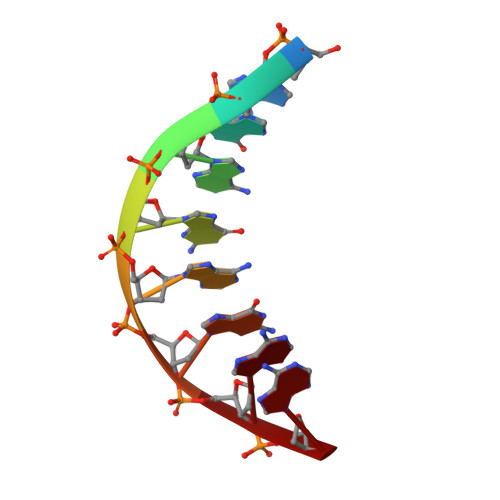
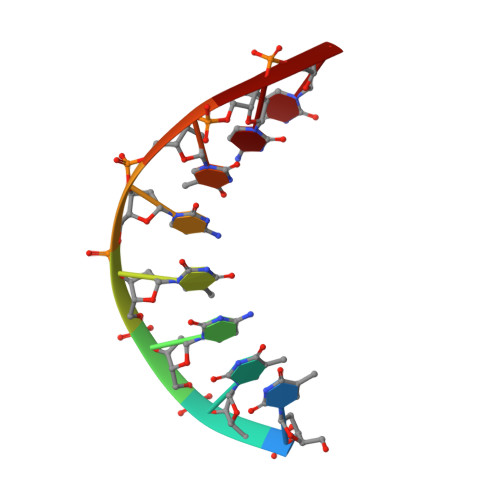
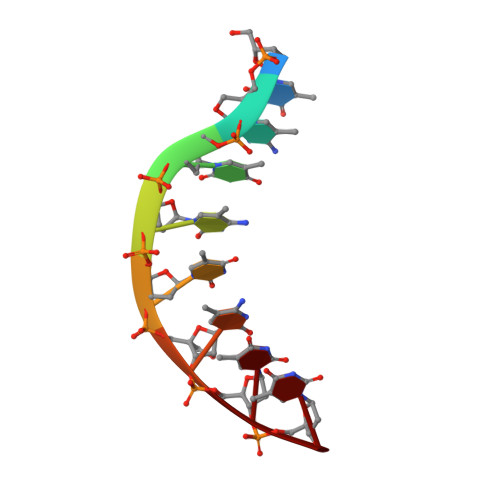
Solution Structure of a DsDNA:Lna Triplex
Sorensen, J.J., Nielsen, J.T., Petersen, M.(2004) Nucleic Acids Res 32: 6078
- PubMed: 15550567
- DOI: https://doi.org/10.1093/nar/gkh942
- Primary Citation of Related Structures:
1W86 - PubMed Abstract:
We have determined the NMR structure of an intramolecular dsDNA:LNA triplex, where the LNA strand is composed of alternating LNA and DNA nucleotides. The LNA oligonucleotide binds to the dsDNA duplex in the major groove by formation of Hoogsteen hydrogen bonds to the purine strand of the duplex. The structure of the dsDNA duplex is changed to accommodate the LNA strand, and it adopts a geometry intermediate between A- and B-type. There is a substantial propeller twist between base-paired nucleobases. This propeller twist and a concomitant large propeller twist between the purine and LNA strands allows the pyrimidines of the LNA strand to interact with the 5'-flanking duplex pyrimidines. Altogether, the triplex has a regular global geometry as shown by a straight helix axis. This shows that even though the third strand is composed of alternating DNA and LNA monomers with different sugar puckers, it forms a seamless triplex. The thermostability of the triplex is increased by 19 degrees C relative to the unmodified DNA triplex at acidic pH. Using NMR spectroscopy, we show that the dsDNA:LNA triplex is stable at pH 8, and that the triplex structure is identical to the structure determined at pH 5.1.
- Nucleic Acid Center, Department of Chemistry, University of Southern Denmark, 5230 Odense M, Denmark.
Organizational Affiliation: