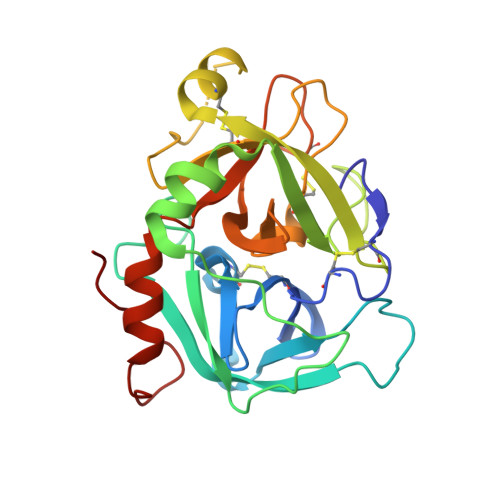
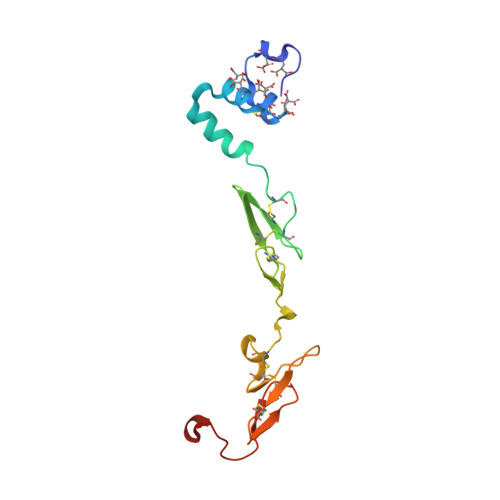
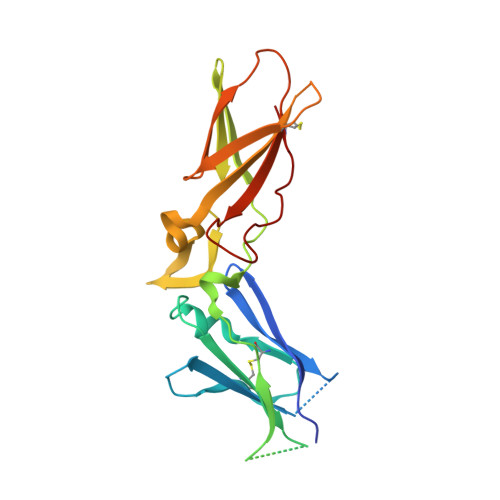
Design of Selective Phenylglycine Amide Tissue Factor/Factor Viia Inhibitors
Groebke-Zbinden, K., Banner, D.W., Ackermann, J., D'Arcy, A., Kirchhofer, D., Ji, Y.-H., Tschopp, T.B., Wallbaum, S., Weber, L.(2005) Bioorg Med Chem Lett 15: 817
- PubMed: 15664864
- DOI: https://doi.org/10.1016/j.bmcl.2004.10.092
- Primary Citation of Related Structures:
1W0Y, 1W2K - PubMed Abstract:
Proof of concept experiments have shown that tissue factor/factor VIIa inhibitors have antithrombotic activity without enhancing bleeding propensity. Starting from lead compounds generated by a biased combinatorial approach, phenylglycine amide tissue factor/factor VIIa inhibitors with low nanomolar affinity and good selectivity against other serine proteases of the coagulation cascade were designed, using the guidance of X-ray structural analysis and molecular modelling.
- F. Hoffmann-La Roche Ltd, CH-4070 Basel, Switzerland. katrin.groebke_zbinden@roche.com
Organizational Affiliation: