Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Chen, Y.-H., Li, M.-H., Zhang, Y., He, L.-L., Yamada, Y., Fitzmaurice, A., Shen, Y., Zhang, H., Tong, L., Yang, J.(2004) Nature 429: 675
- PubMed: 15170217
- DOI: https://doi.org/10.1038/nature02641
- Primary Citation of Related Structures:
1VYT, 1VYU, 1VYV - PubMed Abstract:
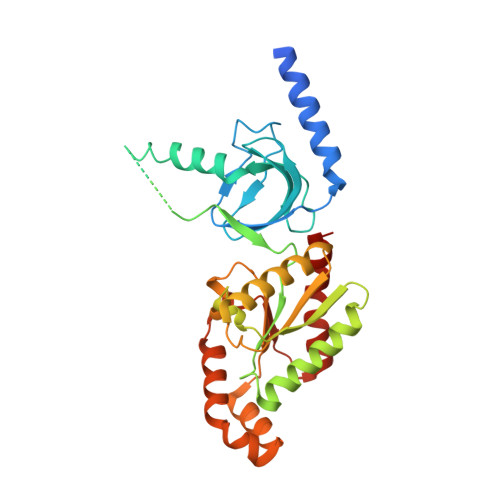
High-voltage-activated Ca2+ channels are essential for diverse biological processes. They are composed of four or five subunits, including alpha1, alpha2-delta, beta and gamma (ref. 1). Their expression and function are critically dependent on the beta-subunit, which transports alpha1 to the surface membrane and regulates diverse channel properties. It is believed that the beta-subunit interacts with alpha1 primarily through the beta-interaction domain (BID), which binds directly to the alpha-interaction domain (AID) of alpha1; however, the molecular mechanism of the alpha1-beta interaction is largely unclear. Here we report the crystal structures of the conserved core region of beta3, alone and in complex with AID, and of beta4 alone. The structures show that the beta-subunit core contains two interacting domains: a Src homology 3 (SH3) domain and a guanylate kinase (GK) domain. The AID binds to a hydrophobic groove in the GK domain through extensive interactions, conferring extremely high affinity between alpha1 and beta-subunits. The BID is essential both for the structural integrity of and for bridging the SH3 and GK domains, but it does not participate directly in binding alpha1. The presence of multiple protein-interacting modules in the beta-subunit opens a new dimension to its function as a multi-functional protein.
- Department of Biological Sciences, Columbia University, New York, New York 10027, USA.
Organizational Affiliation: