Co-Crystal Structure of the HNF-3/Fork Head DNA-Recognition Motif Resembles Histone H5
Clark, K.L., Halay, E.D., Lai, E., Burley, S.K.(1993) Nature 364: 412-420
- PubMed: 8332212
- DOI: https://doi.org/10.1038/364412a0
- Primary Citation of Related Structures:
1VTN - PubMed Abstract:
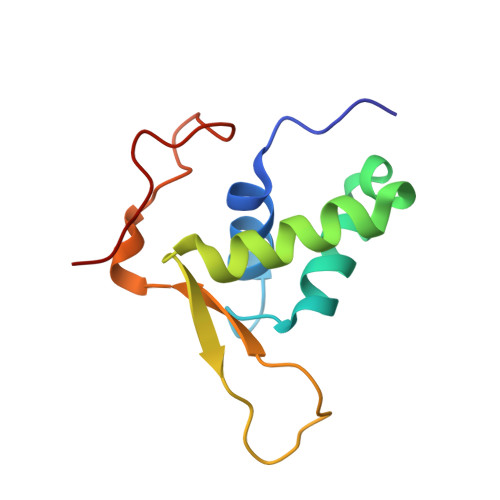
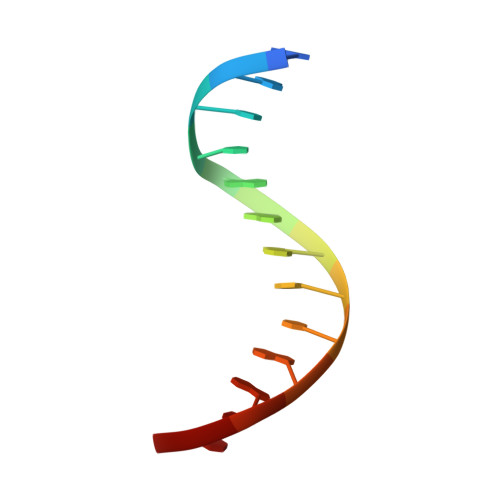
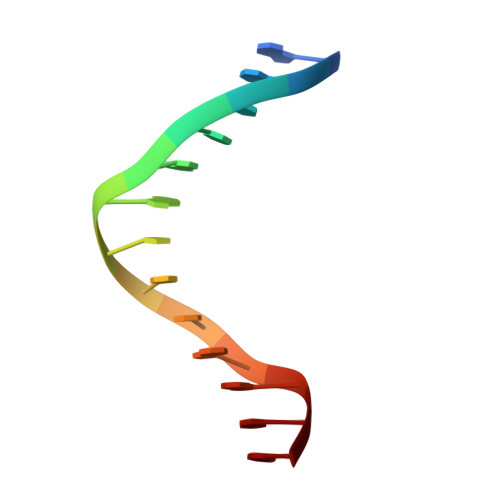
The three-dimensional structure of an HNF-3/fork head DNA-recognition motif complexed with DNA has been determined by X-ray crystallography at 2.5 A resolution. This alpha/beta protein binds B-DNA as a monomer, through interactions with the DNA backbone and through both direct and water-mediated major and minor groove base contacts, inducing a 13 degrees bend. The transcription factor fold is very similar to the structure of histone H5. In its amino-terminal half, three alpha-helices adopt a compact structure that presents the third helix to the major groove. The remainder of the protein includes a twisted, antiparallel beta-structure and random coil that interacts with the minor groove.
- Laboratory of Molecular Biophysics, Rockefeller University, New York, New York 10021.
Organizational Affiliation: