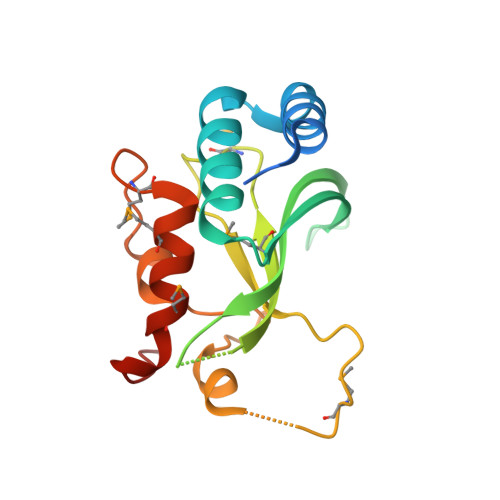
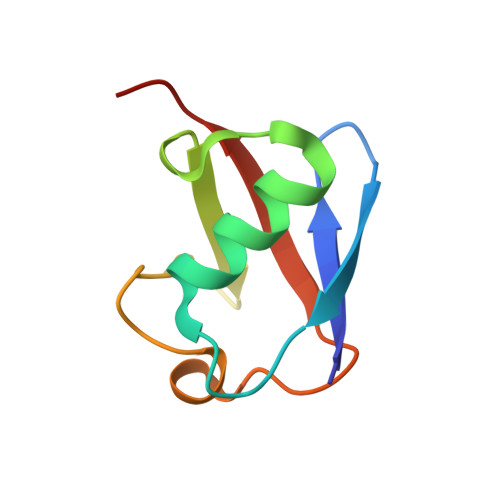
Structural Insights Into Endosomal Sorting Complex Required for Transport (Escrt-I) Recognition of Ubiquitinated Proteins
Teo, H., Veprintsev, D., Williams, R.L.(2004) J Biological Chem 279: 28689
- PubMed: 15044434
- DOI: https://doi.org/10.1074/jbc.M400023200
- Primary Citation of Related Structures:
1UZX - PubMed Abstract:
The endosomal sorting complex required for transport (ESCRT-I) is a 350-kDa complex of three proteins, Vps23, Vps28, and Vps37. The N-terminal ubiquitin-conjugating enzyme E2 variant (UEV) domain of Vps23 is required for sorting ubiquitinated proteins into the internal vesicles of multivesicular bodies. UEVs are homologous to E2 ubiquitin ligases but lack the conserved cysteine residue required for catalytic activity. The crystal structure of the yeast Vps23 UEV in a complex with ubiquitin (Ub) shows the detailed interactions made with the bound Ub. Compared with the solution structure of the Tsg101 UEV (the human homologue of Vps23) in the absence of Ub, two loops that are conserved among the ESCRT-I UEVs move toward each other to grip the Ub in a pincer-like grasp. The contacts with the UEV encompass two adjacent patches on the surface of the Ub, one containing several hydrophobic residues, including Ile-8(Ub), Ile-44(Ub), and Val-70(Ub), and the second containing a hydrophilic patch including residues Asn-60(Ub), Gln-62(Ub), Glu-64(Ub). The hydrophobic Ub patch interacting with the Vps23 UEV overlaps the surface of Ub interacting with the Vps27 ubiquitin-interacting motif, suggesting a sequential model for ubiquitinated cargo binding by these proteins. In contrast, the hydrophilic patch encompasses residues uniquely interacting with the ESCRT-I UEV. The structure provides a detailed framework for design of mutants that can specifically affect ESCRT-I-dependent sorting of ubiquitinated cargo without affecting Vps27-mediated delivery of cargo to endosomes.
- Medical Research Council Laboratory of Molecular Biology and Centre for Protein Engineering, Medical Research Council Centre, Cambridge CB2 2QH, United Kingdom.
Organizational Affiliation: