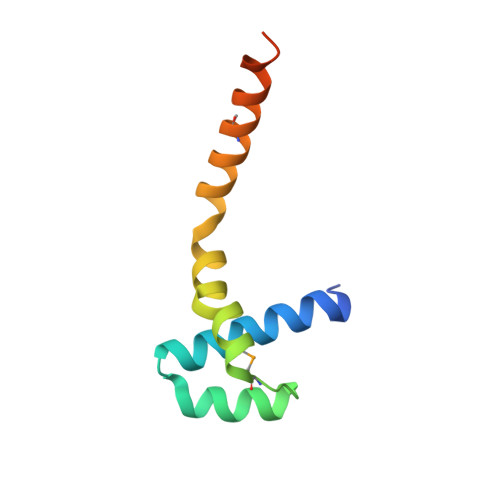
The Structure of the N-Terminal Domain of the Product of the Lissencephaly Gene Lis1 and its Functional Implications
Kim, M.H., Cooper, D.R., Oleksy, A., Devedjiev, Y., Derewenda, U., Reiner, O., Otlewski, J., Derewenda, Z.S.(2004) Structure 12: 987
- PubMed: 15274919
- DOI: https://doi.org/10.1016/j.str.2004.03.024
- Primary Citation of Related Structures:
1UUJ - PubMed Abstract:
Mutations in the Lis1 gene result in lissencephaly (smooth brain), a debilitating developmental syndrome caused by the impaired ability of postmitotic neurons to migrate to their correct destination in the cerebral cortex. Sequence similarities suggest that the LIS1 protein contains a C-terminal seven-blade beta-propeller domain, while the structure of the N-terminal fragment includes the LisH (Lis-homology) motif, a pattern found in over 100 eukaryotic proteins with a hitherto unknown function. We present the 1.75 A resolution crystal structure of the N-terminal domain of mouse LIS1, and we show that the LisH motif is a novel, thermodynamically very stable dimerization domain. The structure explains the molecular basis of a low severity form of lissencephaly.
- Department of Molecular Physiology and Biological Physics and Cancer Center, University of Virginia, Charlottesville, VA 22908, USA.
Organizational Affiliation: