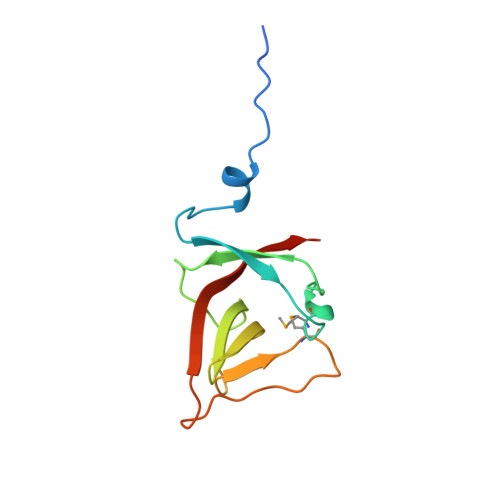
Structure of the UmuD' protein and its regulation in response to DNA damage.
Peat, T.S., Frank, E.G., McDonald, J.P., Levine, A.S., Woodgate, R., Hendrickson, W.A.(1996) Nature 380: 727-730
- PubMed: 8614470
- DOI: https://doi.org/10.1038/380727a0
- Primary Citation of Related Structures:
1UMU - PubMed Abstract:
For life to be sustained, mistakes in DNA repair must be tolerated when damage obscures the genetic information. In bacteria such as Escherichia coli, DNA damage elicits the well regulated 'SOS response'. For the extreme case of damage that cannot be repaired by conventional enzymes, there are proteins that allow the replication of DNA through such lesions, but with a reduction in the fidelity of replication. Essential proteins in this mutagenic process are RecA, DNA polymerase III, UmuD, UmuD' and UmuC (umu: UV mutagenesis). Regulation of this response involves a RecA-mediated self-cleavage of UmuD to produce UmuD'. To understand this system in more detail, we have determined the crystal structure of the E. coli UmuD' mutagenesis protein at 2.5 A resolution. Globular heads folded in an unusual Beta-structure associate to form molecular dimers, and extended amino-terminal tails associate to produce crystallized filaments. The structure provides insight into the mechanism of the self-cleavage reaction that UmuD-like proteins undergo as part of the global SOS response.
- Deparment of Biochemistry and Molecular Biophysics, Columbia University, NY 10032, USA.
Organizational Affiliation: