Human alpha-thrombin inhibition by the active site titrant N alpha-(N,N-dimethylcarbamoyl)-alpha-azalysine p-nitrophenyl ester: a comparative kinetic and X-ray crystallographic study.
Nardini, M., Pesce, A., Rizzi, M., Casale, E., Ferraccioli, R., Balliano, G., Milla, P., Ascenzi, P., Bolognesi, M.(1996) J Mol Biology 258: 851-859
- PubMed: 8637015
- DOI: https://doi.org/10.1006/jmbi.1996.0292
- Primary Citation of Related Structures:
1UMA - PubMed Abstract:
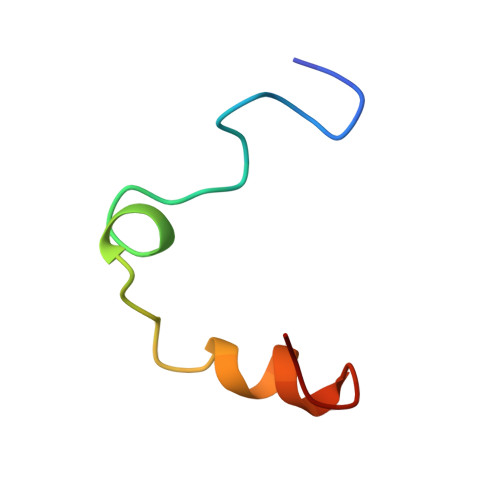
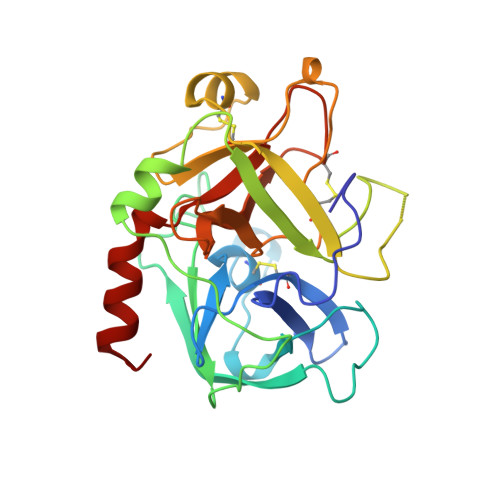
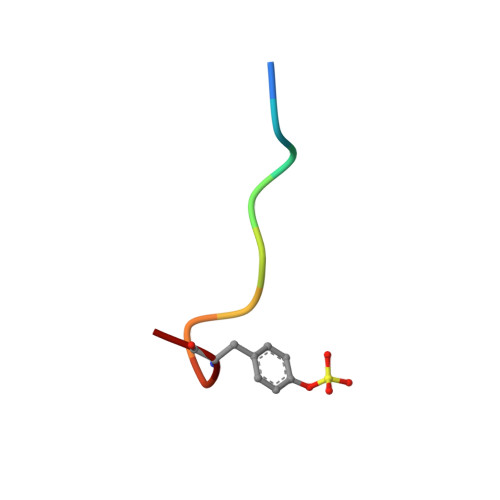
Kinetics for the hydrolysis of the chromogenic active site titrant N alpha-(N,N-dimethylcarbamoyl)-alpha-azalysine p-nitrophenyl ester (Dmc-azaLys-ONp) catalyzed by bovine beta-trypsin, bovine alpha-thrombin, human alpha-thrombin, human Lys77-plasmin, human urinary kallikrein, the M(r) 33,000 and M(r) 54,000 species of human urokinase, as well as by porcine pancreatic beta-kallikrein-A and B have been obtained between pH 6.0 and 8.0, at 21.0 degrees C. Moreover, the three dimensional structure of the human alpha-thrombin-(hirugen).Dmc-azaLys acyl.enzyme complex has been analyzed and refined by X-ray crystallography at 2.0 A resolution (R-factor = 0.168). As observed for bovine beta-trypsin, the acylating inhibitor molecule is covalently bound to the Ser195 catalytic residue, filling the human alpha-thrombin S1 primary specificity subsite with its lysyl side-group. However, the carbonyl group of the scissile human alpha-thrombin.Dmc-azaLys acyl bond does not occupy properly the oxyanion binding hole. At variance from the bovine beta-trypsin.Dmc-azaLys acyl.enzyme structure, a second, not covalently bound, inhibitor molecule, partly shielded by the 60-insertion loop of human alpha-thrombin, is contacting the enzyme "aryl-binding site".
- Centro Biotecnologie Avanzate IST, Università di Genova, Italy.
Organizational Affiliation: