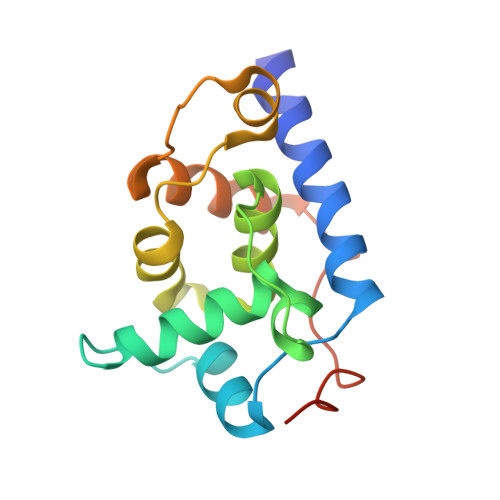
Conformational switching by the scaffolding protein D directs the assembly of bacteriophage phiX174
Morais, M.C., Fisher, M., Kanamaru, S., Przybyla, L., Burgner, J., Fane, B.A., Rossmann, M.G.(2004) Mol Cell 15: 991-997
- PubMed: 15383287
- DOI: https://doi.org/10.1016/j.molcel.2004.08.023
- Primary Citation of Related Structures:
1TX9 - PubMed Abstract:
The three-dimensional structure of bacteriophage phiX174 external scaffolding protein D, prior to its interaction with other structural proteins, has been determined to 3.3 angstroms by X-ray crystallography. The crystals belong to space group P4(1)2(1)2 with a dimer in the asymmetric unit that closely resembles asymmetric dimers observed in the phiX174 procapsid structure. Furthermore, application of the crystallographic 4(1) symmetry operation to one of these dimers generates a tetramer similar to the tetramer in the icosahedral asymmetric unit of the procapsid. These data suggest that both dimers and tetramers of the D protein are true morphogenetic intermediates and can form independently of other proteins involved in procapsid morphogenesis. The crystal structure of the D scaffolding protein thus represents the state of the polypeptide prior to procapsid assembly. Hence, comparison with the procapsid structure provides a rare opportunity to follow the conformational switching events necessary for the construction of complex macromolecular assemblies.
- Department of Biological Sciences, Purdue University, 915 West State Street, West Lafayette, IN 47907, USA.
Organizational Affiliation: