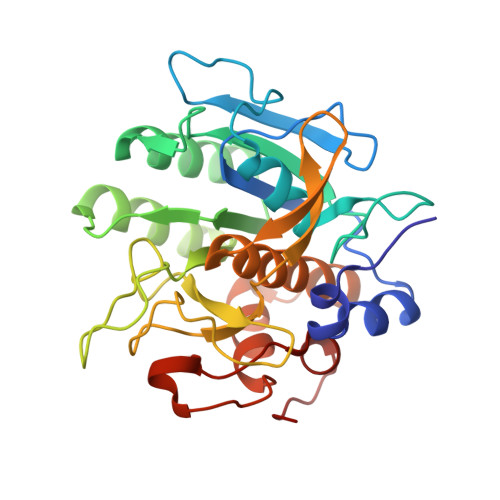
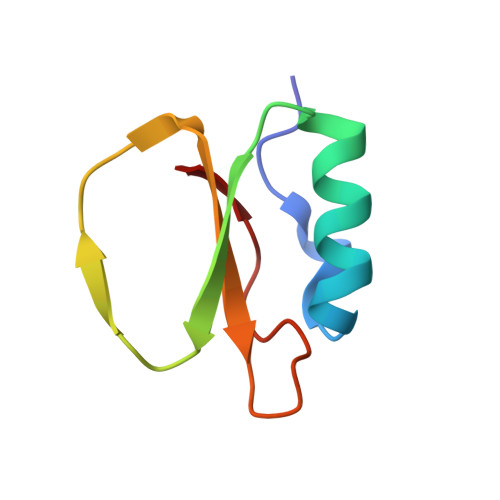
Binding, Proteolytic, and Crystallographic Analyses of Mutations at the Protease-Inhibitor Interface of the Subtilisin BPN'/Chymotrypsin Inhibitor 2 Complex(,).
Radisky, E.S., Kwan, G., Karen Lu, C.J., Koshland Jr., D.E.(2004) Biochemistry 43: 13648-13656
- PubMed: 15504027
- DOI: https://doi.org/10.1021/bi048797k
- Primary Citation of Related Structures:
1TM1, 1TM3, 1TM4, 1TM5, 1TM7, 1TMG, 1TO1, 1TO2 - PubMed Abstract:
A series of mutants of chymotrypsin inhibitor 2 (CI2), at residues that interact with the inhibited enzyme subtilisin BPN', were studied to determine the relative importance of intermolecular contacts on either side of the scissile bond. Mutants were tested for inhibition of subtilisin, rates of hydrolysis by subtilisin, and ability to acylate subtilisin. Additionally, crystal structures of the mutant CI2 complexes with subtilisin were obtained. Ordered water molecules were found to play an important role in inhibitor recognition, and features of the crystal structures, in combination with biochemical data, support a transition-state stabilization role for the P(1) residue in subtilisin catalysis. Consistent with the proposed mechanism of inhibition, in which rapid acylation is followed by religation, leaving-group contacts with the enzyme were found to be more critical determinants of inhibition than acylating-group contacts in the mutants studied here.
- Department of Molecular and Cell Biology, University of California, Berkeley, California 94720, USA.
Organizational Affiliation: