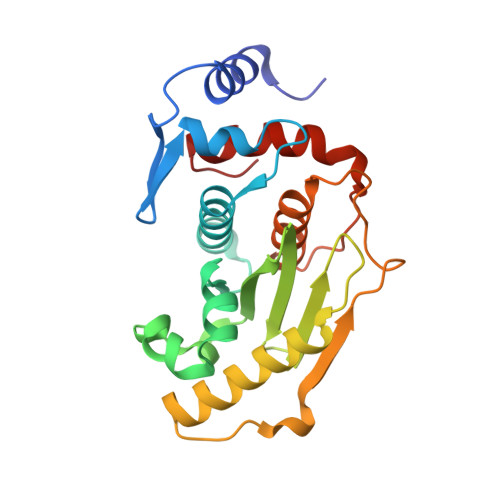
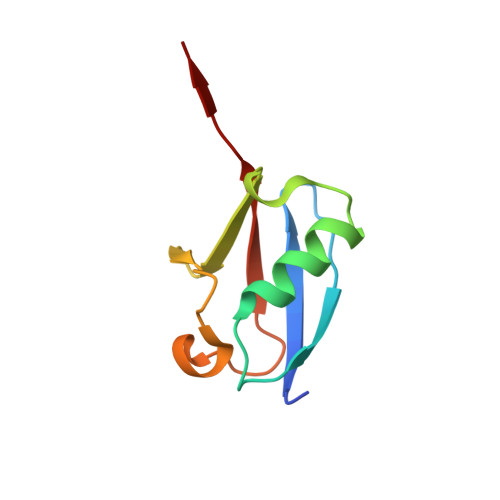
A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2-SUMO complex
Reverter, D., Lima, C.D.(2004) Structure 12: 1519-1531
- PubMed: 15296745
- DOI: https://doi.org/10.1016/j.str.2004.05.023
- Primary Citation of Related Structures:
1TGZ, 1TH0 - PubMed Abstract:
Modification of cellular proteins by the ubiquitin-like protein SUMO is essential for nuclear metabolism and cell cycle progression in yeast. X-ray structures of the human Senp2 catalytic protease domain and of a covalent thiohemiacetal transition-state complex obtained between the Senp2 catalytic domain and SUMO-1 revealed details of the respective protease and substrate surfaces utilized in interactions between these two proteins. Comparative biochemical and structural analysis between Senp2 and the yeast SUMO protease Ulp1 revealed differential abilities to process SUMO-1, SUMO-2, and SUMO-3 in maturation and deconjugation reactions. Further biochemical characterization of the three SUMO isoforms into which an additional Gly-Gly di-peptide was inserted, or whereby the respective SUMO tails from the three isoforms were swapped, suggests a strict dependence for SUMO isopeptidase activity on residues C-terminal to the conserved Gly-Gly motif and preferred cleavage site for SUMO proteases.
- Structural Biology Program, Sloan-Kettering Institute, New York, New York 10021, USA.
Organizational Affiliation: