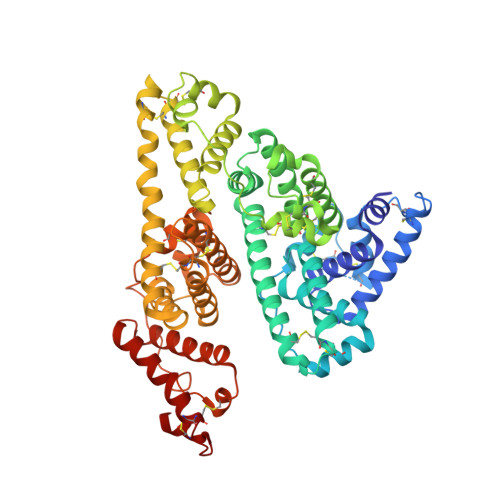
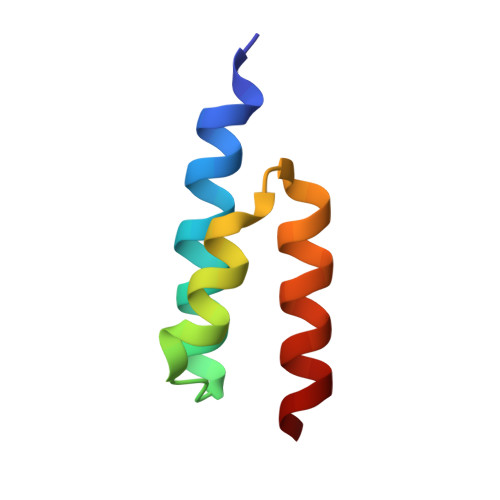
Crystal structure and biological implications of a bacterial albumin binding module in complex with human serum albumin
Lejon, S., Frick, I.-M., Bjorck, L., Wikstrom, M., Svensson, S.(2004) J Biological Chem 279: 42924-42928
- PubMed: 15269208
- DOI: https://doi.org/10.1074/jbc.M406957200
- Primary Citation of Related Structures:
1TF0 - PubMed Abstract:
Many bactericide species express surface proteins that interact with human serum albumin (HSA). Protein PAB from the anaerobic bacterium Finegoldia magna (formerly Peptostreptococcus magnus) represents one of these proteins. Protein PAB contains a domain of 53 amino acid residues known as the GA module. GA homologs are also found in protein G of group C and G streptococci. Here we report the crystal structure of HSA in complex with the GA module of protein PAB. The model of the complex was refined to a resolution of 2.7 A and reveals a novel binding epitope located in domain II of the albumin molecule. The GA module is composed of a left-handed three-helix bundle, and residues from the second helix and the loops surrounding it were found to be involved in HSA binding. Furthermore, the presence of HSA-bound fatty acids seems to influence HSA-GA complex formation. F. magna has a much more restricted host specificity compared with C and G streptococci, which is also reflected in the binding of different animal albumins by proteins PAB and G. The structure of the HSA-GA complex offers a molecular explanation to this unusually clear example of bacterial adaptation.
- Department of Cell and Molecular Biology, Uppsala University, SE-751 24 Uppsala, Sweden. sara@xray.bmc.uu.se
Organizational Affiliation: