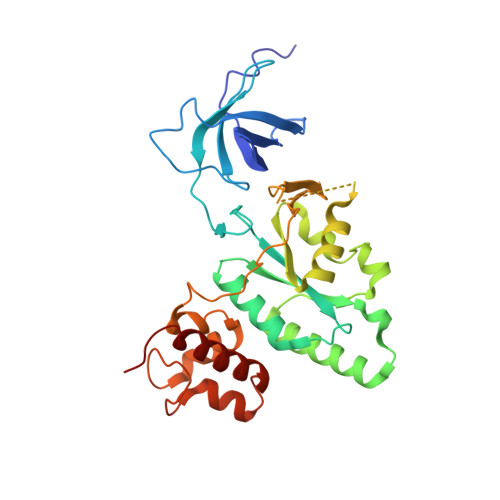
The Crystal Structure of YloQ, a Circularly Permuted GTPase Essential for Bacillus Subtilis Viability.
Levdikov, V.M., Blagova, E.V., Brannigan, J.A., Cladiere, L., Antson, A.A., Isupov, M.N., Seror, S.J., Wilkinson, A.J.(2004) J Mol Biology 340: 767-782
- PubMed: 15223319
- DOI: https://doi.org/10.1016/j.jmb.2004.05.029
- Primary Citation of Related Structures:
1T9H - PubMed Abstract:
yloQ is one of 11 essential genes in Bacillus subtilis with unknown roles in the physiology of the cell. It encodes a polypeptide of 298 residues with motifs characteristic of GTPases. As a contribution to elucidating its indispensable cellular function, we have solved the crystal structure of YloQ to 1.6 A spacing, revealing a three-domain organisation. At the heart of the molecule is the putative GTPase domain, which exhibits a classical alpha/beta nucleotide-binding fold with a topology very similar to that of Ras and Era. However, as anticipated from the order in which the conserved G protein motifs appear in the sequence, the GTPase domain fold in YloQ is circularly permuted with respect to the classical GTPases. The nucleotide-binding pocket in YloQ is unoccupied, and analysis of the phosphate-binding (P) loop indicates that conformational changes in this region would be needed to accommodate GTP. The GTPase domain is flanked at its N terminus by a beta-barrel domain with an oligonucleotide/oligosaccharide-binding (OB) fold, and at its C terminus by an alpha-helical domain containing a coordinated zinc ion. This combination of protein modules is unique to YloQ and its orthologues. Sequence comparisons reveal a clustering of conserved basic and aromatic residues on one face of the OB domain, perhaps pointing to a role for YloQ in nucleic acid binding. The zinc ion in the alpha-helical domain is coordinated by three cysteine residues and a histidine residue in a novel ligand organisation. The juxtaposition of the switch I and switch II regions of the G domain and the OB and zinc-binding domains suggests that chemical events at the GTPase active site may be transduced into relative movements of these domains. The pattern of conserved residues and electrostatic surface potential calculations suggest that the OB and/or Zn-binding domains participate in nucleic acid binding consistent with a possible role for YloQ at some stage during mRNA translation.
- Structural Biology Laboratory, Department of Chemistry, University of York, York YO10 5YW, UK.
Organizational Affiliation: