Oxyguanidines. Part 2: Discovery of a novel orally active thrombin inhibitor through structure-based drug design and parallel synthesis
Lu, T., Markotan, T., Coppo, F., Tomczuk, B., Crysler, C., Eisennagel, S., Spurlino, J., Gremminger, L., Soll, R.M., Giardino, E.C., Bone, R.(2004) Bioorg Med Chem Lett 14: 3727-3731
- PubMed: 15203151
- DOI: https://doi.org/10.1016/j.bmcl.2004.05.002
- Primary Citation of Related Structures:
1T4U, 1T4V - PubMed Abstract:
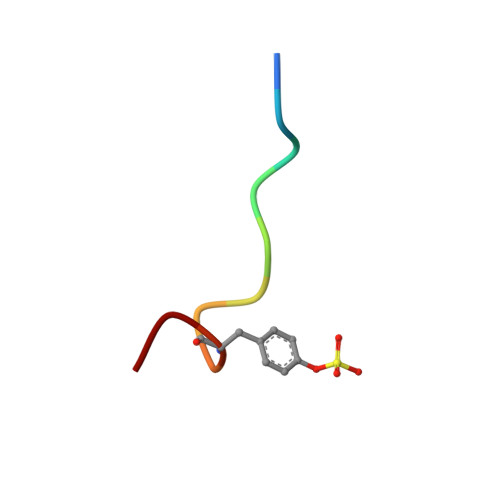
Through structure-based drug design and parallel synthesis, we have discovered a novel series of nonpeptidic phenyl-based thrombin inhibitors using oxyguanidines as guanidine bioisosteres. These compounds have been found to be highly potent, highly selective, and orally bioavailable.
- 3-Dimensional Pharmaceuticals, Inc., 665 Stockton Drive, Exton, PA 19341, USA. tlu3@prdus.jnj.com
Organizational Affiliation: