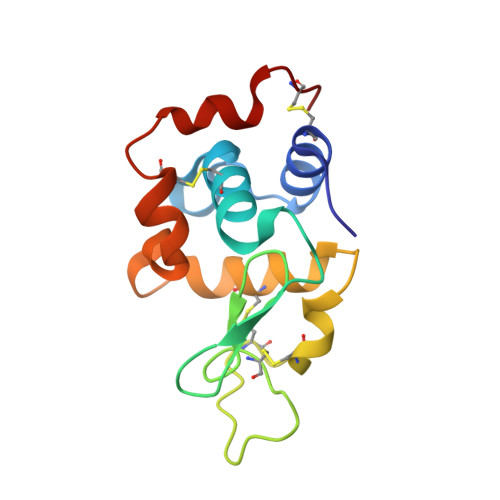
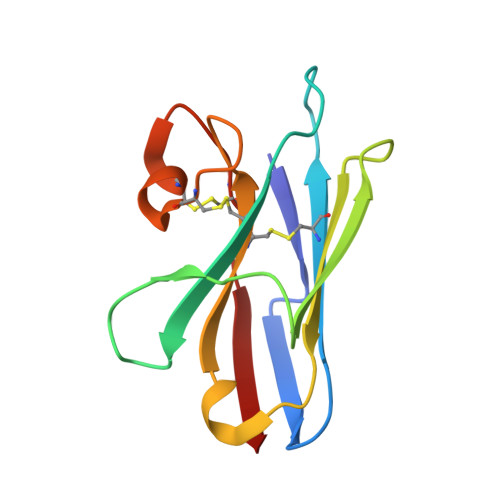
Crystal structure of a shark single-domain antibody V region in complex with lysozyme.
Stanfield, R.L., Dooley, H., Flajnik, M.F., Wilson, I.A.(2004) Science 305: 1770-1773
- PubMed: 15319492
- DOI: https://doi.org/10.1126/science.1101148
- Primary Citation of Related Structures:
1SQ2, 1T6V - PubMed Abstract:
Cartilaginous fish are the phylogenetically oldest living organisms known to possess components of the vertebrate adaptive immune system. Key to their immune response are heavy-chain, homodimeric immunoglobulins called new antigen receptors (IgNARs), in which the variable (V) domains recognize antigens with only a single immunoglobulin domain, akin to camelid heavy-chain V domains. The 1.45 angstrom resolution crystal structure of the type I IgNAR V domain in complex with hen egg-white lysozyme (HEL) reveals a minimal antigen-binding domain that contains only two of the three conventional complementarity-determining regions but still binds HEL with nanomolar affinity by means of a binding interface comparable in size to conventional antibodies.
- Department of Molecular Biology, Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.
Organizational Affiliation: