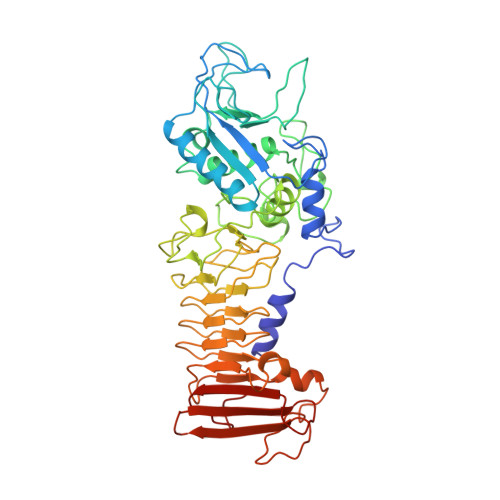
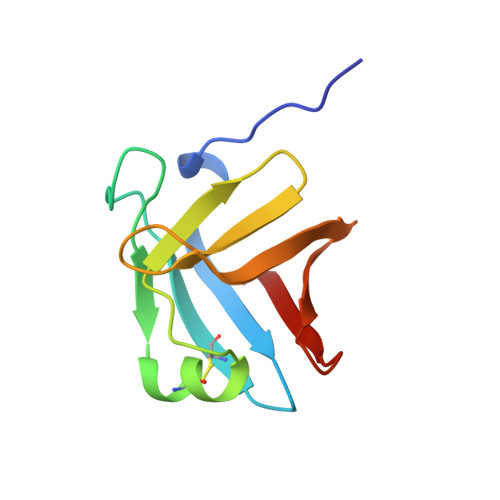
Crystal structure of a complex between Serratia marcescens metallo-protease and an inhibitor from Erwinia chrysanthemi.
Baumann, U., Bauer, M., Letoffe, S., Delepelaire, P., Wandersman, C.(1995) J Mol Biology 248: 653-661
- PubMed: 7752231
- DOI: https://doi.org/10.1006/jmbi.1995.0249
- Primary Citation of Related Structures:
1SMP - PubMed Abstract:
The crystal structure of the complex between the 50 kDa metallo-endoproteinase from Serratia marcescens (SMP), a member of the metzincin superfamily, and an inhibitor from Erwinia chrysanthemi (Inh) was solved by molecular replacement using the known structure of SMP, and refined at 2.30 A resolution to a crystallographic R-factor of 0.195. The E. chrysanthemi inhibitor folds into a compact eight-stranded antiparallel beta-barrel of simple up-down topology such as is found for members of the retinol binding protein family. It mainly interacts with the protease via its five N-terminal residues, which insert into the active site cleft, occupying the S' sites. The first N-terminal residue, SerI1, is partially cleaved off by the protease, while SerI2 makes a hydrogen bond with the catalytically active glutamic acid, Glu177, of the protease. Further interactions are made between one face of the inhibitor formed by the strands s3, s4 and s5 and the protease segment 218 to 228, which is located immediately after the characteristic "Met-turn" of the metzincins.
- Institut für Organische Chemie und Biochemie der Universität, Freiburg, Germany.
Organizational Affiliation: