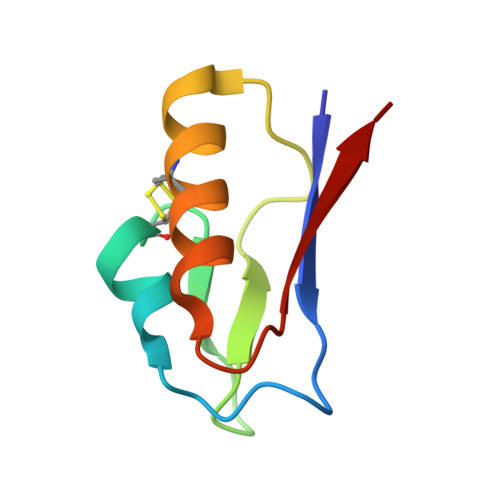
The 1.5 A Resolution Crystal Structure of [Fe3S4]-Ferredoxin from the Hyperthermophilic Archaeon Pyrococcus furiosus
Nielsen, M.S., Harris, P., Ooi, B.L., Christensen, H.E.M.(2004) Biochemistry 43: 5188-5194
- PubMed: 15122884
- DOI: https://doi.org/10.1021/bi049942x
- Primary Citation of Related Structures:
1SIZ, 1SJ1 - PubMed Abstract:
The structure of [Fe(3)S(4)]-ferredoxin from the hyperthermophilic archaeon Pyrococcus furiosus has been determined to 1.5 A resolution from a crystal belonging to space group P2(1) with two molecules in the asymmetric unit. The structure has been solved with molecular replacement by use of the ferredoxin from Thermotoga maritima. The fold is similar to that of related monocluster ferredoxins and contains two double-stranded antiparallel beta-sheets and two alpha-helices. The hydrophobic interaction between Trp2 and Tyr46 is confirmed, linking the C-terminus to the longer alpha-helix. The structure contains a double-conformation disulfide bond existing in a left-handed and a right-handed spiral conformation. The crystal packing reveals a beta-sheet interaction, which supports the suggestion that P. furiosus ferredoxin is a functional dimer. The extraordinary thermostability of P. furiosus ferredoxin is further discussed.
- Department of Chemistry, Building 207, Technical University of Denmark, DK-2800 Kgs. Lyngby, Denmark.
Organizational Affiliation: