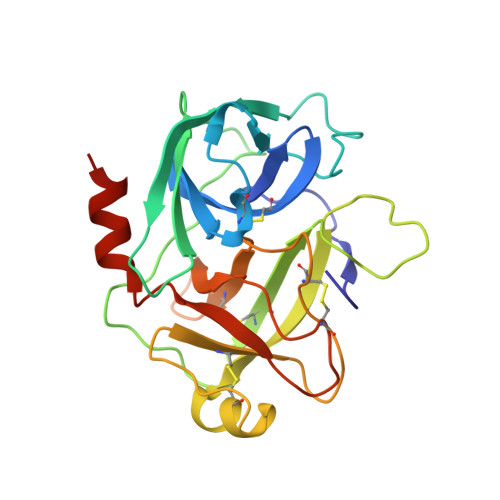
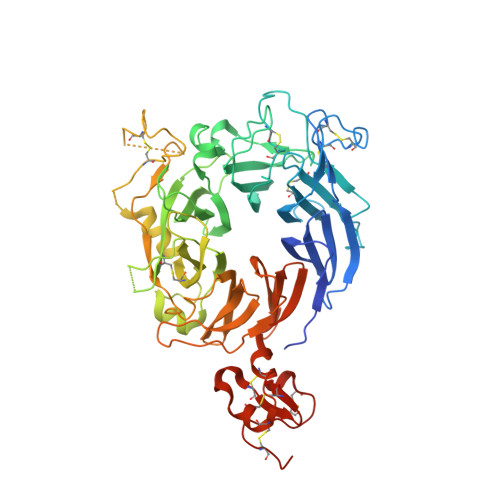
Crystal structure of the HGF beta-chain in complex with the Sema domain of the Met receptor.
Stamos, J., Lazarus, R.A., Yao, X., Kirchhofer, D., Wiesmann, C.(2004) EMBO J 23: 2325-2335
- PubMed: 15167892
- DOI: https://doi.org/10.1038/sj.emboj.7600243
- Primary Citation of Related Structures:
1SHY - PubMed Abstract:
The Met tyrosine kinase receptor and its ligand, hepatocyte growth factor (HGF), play important roles in normal development and in tumor growth and metastasis. HGF-dependent signaling requires proteolysis from an inactive single-chain precursor into an active alpha/beta-heterodimer. We show that the serine protease-like HGF beta-chain alone binds Met, and report its crystal structure in complex with the Sema and PSI domain of the Met receptor. The Met Sema domain folds into a seven-bladed beta-propeller, where the bottom face of blades 2 and 3 binds to the HGF beta-chain 'active site region'. Mutation of HGF residues in the area that constitutes the active site region in related serine proteases significantly impairs HGF beta binding to Met. Key binding loops in this interface undergo conformational rearrangements upon maturation and explain the necessity of proteolytic cleavage for proper HGF signaling. A crystallographic dimer interface between two HGF beta-chains brings two HGF beta:Met complexes together, suggesting a possible mechanism of Met receptor dimerization and activation by HGF.
- Department of Protein Engineering, Genentech Inc., South San Francisco, CA 94080, USA.
Organizational Affiliation: