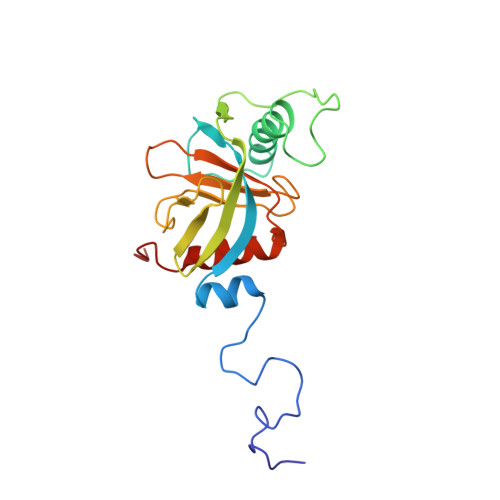
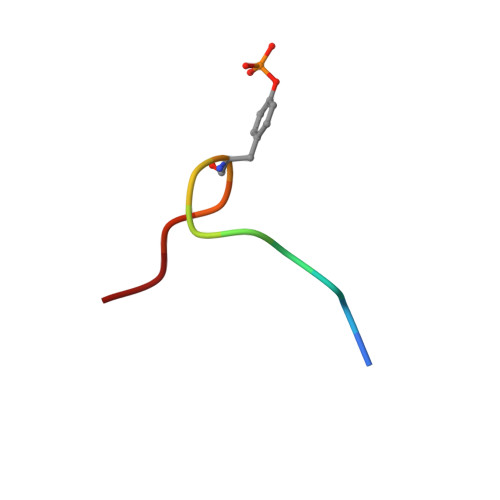
Structure and ligand recognition of the phosphotyrosine binding domain of Shc.
Zhou, M.M., Ravichandran, K.S., Olejniczak, E.F., Petros, A.M., Meadows, R.P., Sattler, M., Harlan, J.E., Wade, W.S., Burakoff, S.J., Fesik, S.W.(1995) Nature 378: 584-592
- PubMed: 8524391
- DOI: https://doi.org/10.1038/378584a0
- Primary Citation of Related Structures:
1SHC - PubMed Abstract:
The nuclear magnetic resonance structure of the phosphotyrosine binding (PTB) domain of Shc complexed to a phosphopeptide reveals an alternative means of recognizing tyrosine-phosphorylated proteins. Unlike in SH2 domains, the phosphopeptide forms an antiparallel beta-strand with a beta-sheet of the protein, interacts with a hydrophobic pocket through the (pY-5) residue, and adopts a beta-turn. The PTB domain is structurally similar to pleckstrin homology domains (a beta-sandwich capped by an alpha-helix) and binds to acidic phospholipids, suggesting a possible role in membrane localization.
- Pharmaceutical Discovery Division, Abbott Laboratories, Abbott Park, Illinois 60064, USA.
Organizational Affiliation: