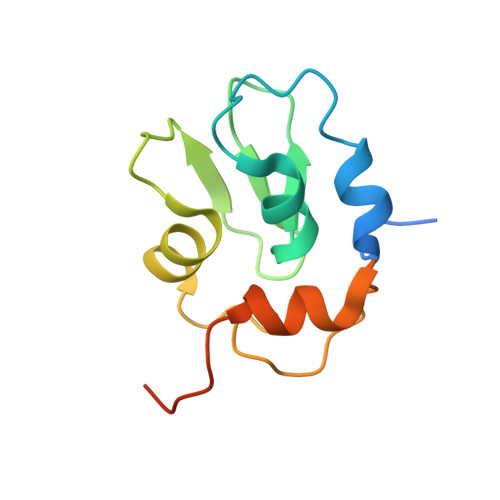
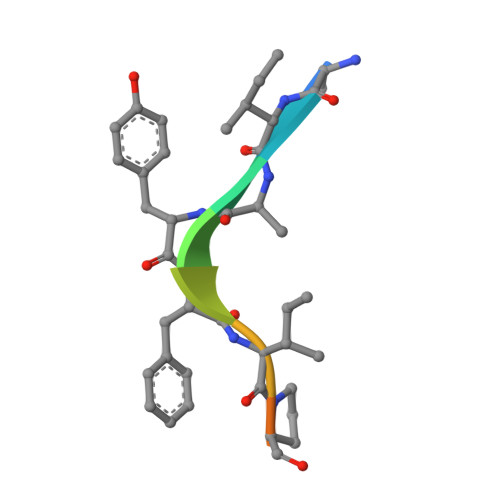
Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim.
Yan, N., Wu, J.W., Chai, J., Li, W., Shi, Y.(2004) Nat Struct Mol Biol 11: 420-428
- PubMed: 15107838
- DOI: https://doi.org/10.1038/nsmb764
- Primary Citation of Related Structures:
1SDZ, 1SE0 - PubMed Abstract:
The Drosophila melanogaster inhibitor of apoptosis protein DIAP1 suppresses apoptosis in part through inhibition of the effector caspase DrICE. The pro-death proteins Reaper, Hid and Grim (RHG) induce apoptosis by antagonizing DIAP1 function. However, the underlying molecular mechanisms remain unknown. Here we demonstrate that DIAP1 directly inhibits the catalytic activity of DrICE through its BIR1 domain and this inhibition is countered effectively by the RHG proteins. Inhibition of DrICE by DIAP1 occurs only after the cleavage of its N-terminal 20 amino acids and involves a conserved surface groove on BIR1. Crystal structures of BIR1 bound to the RHG peptides show that the RHG proteins use their N-terminal IAP-binding motifs to bind to the same surface groove, hence relieving DIAP1-mediated inhibition of DrICE. These studies define novel molecular mechanisms for the inhibition and activation of a representative D. melanogaster effector caspase.
- Department of Molecular Biology, Princeton University, Lewis Thomas Laboratory, Washington Road, Princeton, New Jersey 08544, USA.
Organizational Affiliation: