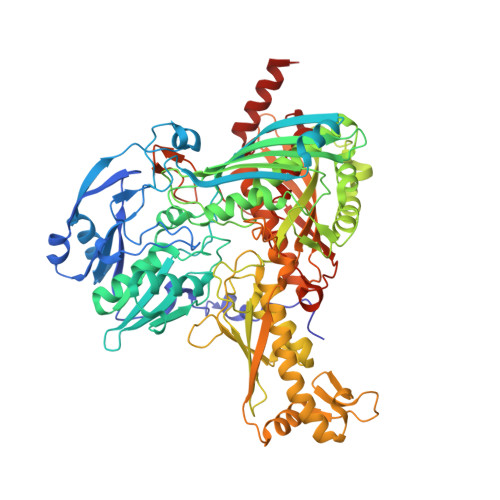
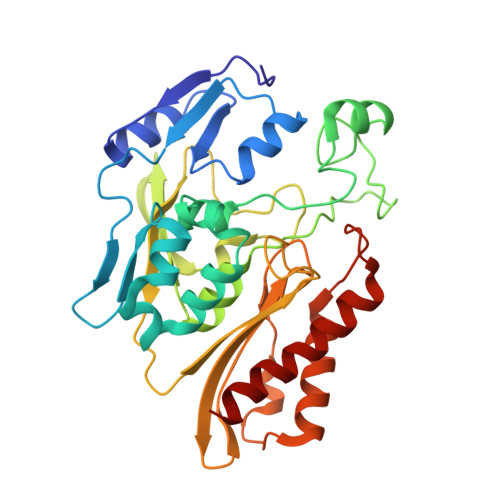
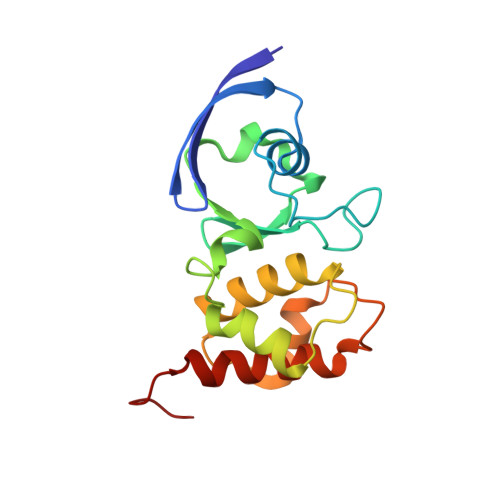
Structure of a Xanthine Oxidase-Related 4-Hydroxybenzoyl-CoA Reductase with an Additional [4Fe-4S] Cluster and an Inverted Electron Flow.
Unciuleac, M., Warkentin, E., Page, C.C., Boll, M., Ermler, U.(2004) Structure 12: 2249-2256
- PubMed: 15576037
- DOI: https://doi.org/10.1016/j.str.2004.10.008
- Primary Citation of Related Structures:
1RM6, 1SB3 - PubMed Abstract:
The Mo-flavo-Fe/S-dependent heterohexameric protein complex 4-hydroxybenzoyl-CoA reductase (4-HBCR, dehydroxylating) is a central enzyme of the anaerobic degradation of phenolic compounds and belongs to the xanthine oxidase (XO) family of molybdenum enzymes. Its X-ray structure was established at 1.6 A resolution. The most pronounced difference between 4-HBCR and other structurally characterized members of the XO family is the insertion of 40 amino acids within the beta subunit, which carries an additional [4Fe-4S] cluster at a distance of 16.5 A to the isoalloxazine ring of FAD. The architecture of 4-HBCR and concomitantly performed electron transfer rate calculations suggest an inverted electron transfer chain from the donor ferredoxin via the [4Fe-4S] cluster to the Mo over a distance of 55 A. The binding site of 4-hydroxybenzoyl-CoA is located in an 18 A long channel lined up by several aromatic side chains around the aromatic moiety, which are proposed to shield and stabilize the postulated radical intermediates during catalysis.
- Institut für Biologie II, Mikrobiologie Schänzlestrasse 1, D-79104 Freiburg, Germany.
Organizational Affiliation: