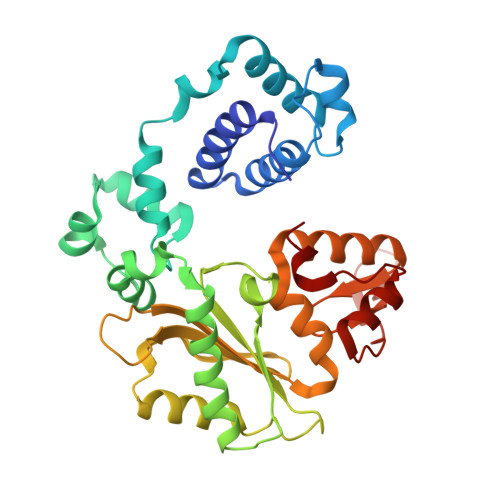
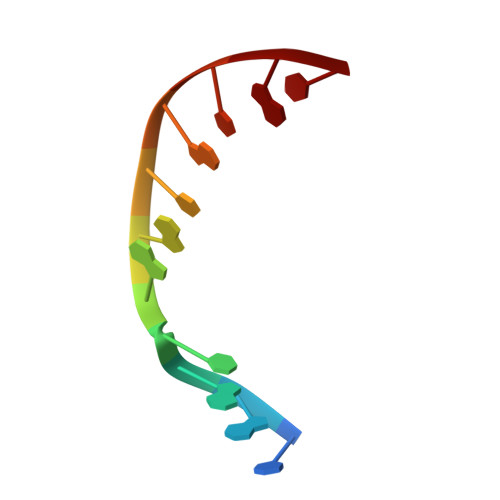
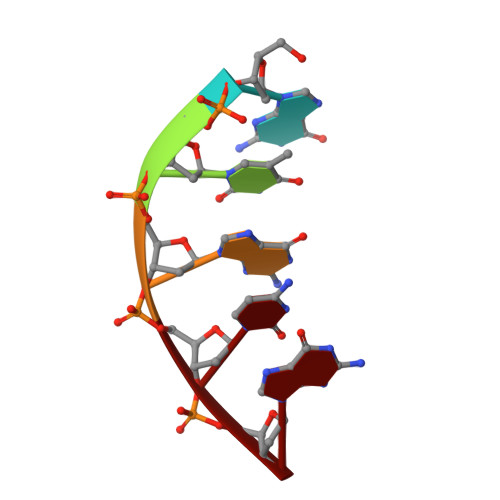
A structural solution for the DNA polymerase lambda-dependent repair of DNA gaps with minimal homology.
Garcia-Diaz, M., Bebenek, K., Krahn, J.M., Blanco, L., Kunkel, T.A., Pedersen, L.C.(2004) Mol Cell 13: 561-572
- PubMed: 14992725
- DOI: https://doi.org/10.1016/s1097-2765(04)00061-9
- Primary Citation of Related Structures:
1RZT - PubMed Abstract:
Human DNA polymerase lambda (Pol lambda) is a family X member with low frameshift fidelity that has been suggested to perform gap-filling DNA synthesis during base excision repair and during repair of broken ends with limited homology. Here, we present a 2.1 A crystal structure of the catalytic core of Pol lambda in complex with DNA containing a two nucleotide gap. Pol lambda makes limited contacts with the template strand at the polymerase active site, and superimposition with Pol beta in a ternary complex suggests a shift in the position of the DNA at the active site that is reminiscent of a deletion intermediate. Surprisingly, Pol lambda can adopt a closed conformation, even in the absence of dNTP binding. These observations have implications for the catalytic mechanism and putative DNA repair functions of Pol lambda.
- Laboratory of Molecular Genetics, National Institute of Environmental Health Sciences, Research Triangle Park, NC 27709, USA.
Organizational Affiliation: