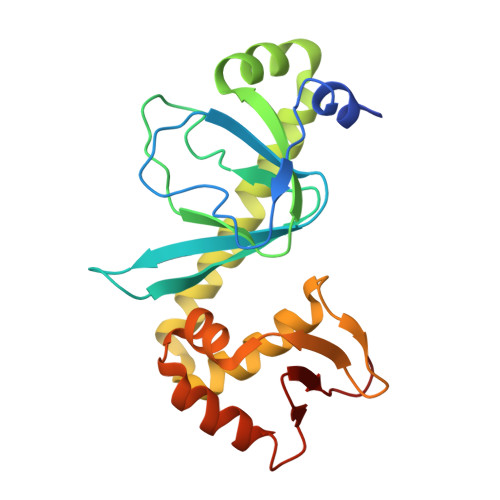
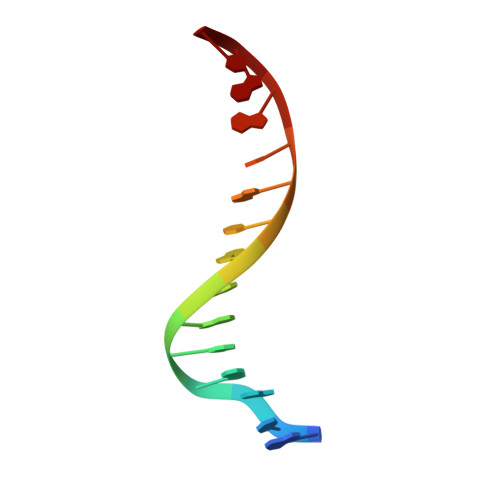
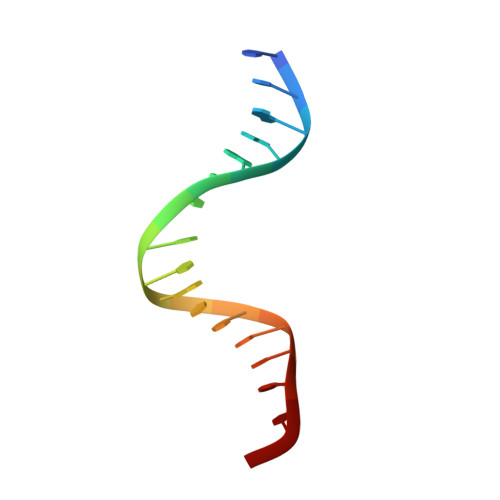
Aromatic hydrogen bond in sequence-specific protein DNA recognition.
Parkinson, G., Gunasekera, A., Vojtechovsky, J., Zhang, X., Kunkel, T.A., Berman, H., Ebright, R.H.(1996) Nat Struct Biol 3: 837-841
- PubMed: 8836098
- DOI: https://doi.org/10.1038/nsb1096-837
- Primary Citation of Related Structures:
1RUN, 1RUO