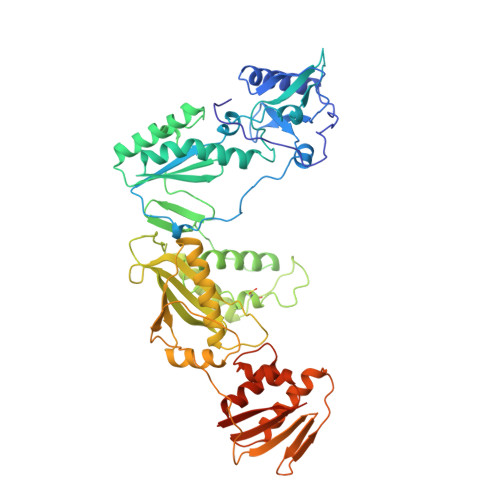
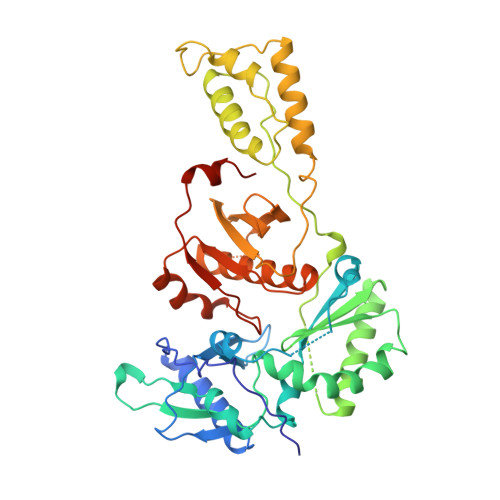
3'-Azido-3'-deoxythymidine drug resistance mutations in HIV-1 reverse transcriptase can induce long range conformational changes.
Ren, J., Esnouf, R.M., Hopkins, A.L., Jones, E.Y., Kirby, I., Keeling, J., Ross, C.K., Larder, B.A., Stuart, D.I., Stammers, D.K.(1998) Proc Natl Acad Sci U S A 95: 9518-9523
- PubMed: 9689112
- DOI: https://doi.org/10.1073/pnas.95.16.9518
- Primary Citation of Related Structures:
1RT3 - PubMed Abstract:
HIV reverse transcriptase (RT) is one of the main targets for the action of anti-AIDS drugs. Many of these drugs [e.g., 3'-azido-3'-deoxythymidine (AZT) and 2',3'-dideoxyinosine (ddI)] are analogues of the nucleoside substrates used by the HIV RT. One of the main problems in anti-HIV therapy is the selection of a mutant virus with reduced drug sensitivity. Drug resistance in HIV is generated for nucleoside analogue inhibitors by mutations in HIV RT. However, most of these mutations are situated some distance from the polymerase active site, giving rise to questions concerning the mechanism of resistance. To understand the possible structural bases for this, the crystal structures of AZT- and ddI-resistant RTs have been determined. For the ddI-resistant RT with a mutation at residue 74, no significant conformational changes were observed for the p66 subunit. In contrast, for the AZT-resistant RT (RTMC) bearing four mutations, two of these (at 215 and 219) give rise to a conformational change that propagates to the active site aspartate residues. Thus, these drug resistance mutations produce an effect at the RT polymerase site mediated simply by the protein. It is likely that such long-range effects could represent a common mechanism for generating drug resistance in other systems.
- Laboratory of Molecular Biophysics, Rex Richards Building, South Parks Road, University of Oxford, Oxford OX1 3QU, United Kingdom.
Organizational Affiliation: