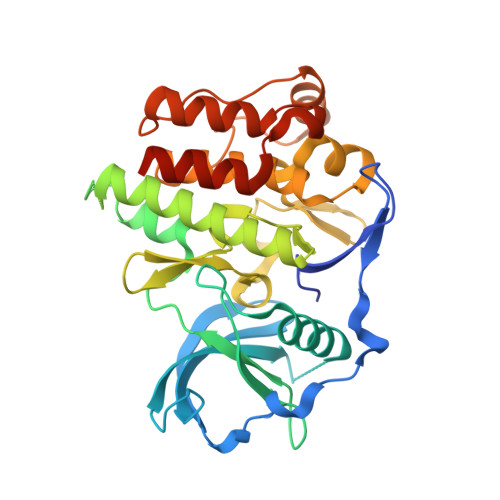
The structural basis for autoinhibition of FLT3 by the juxtamembrane domain.
Griffith, J., Black, J., Faerman, C., Swenson, L., Wynn, M., Lu, F., Lippke, J., Saxena, K.(2004) Mol Cell 13: 169-178
- PubMed: 14759363
- DOI: https://doi.org/10.1016/s1097-2765(03)00505-7
- Primary Citation of Related Structures:
1RJB - PubMed Abstract:
FLT3 is a type III receptor tyrosine kinase that is thought to play a key role in hematopoiesis. Certain classes of FLT3 mutations cause constitutively activated forms of the receptor that are found in significant numbers of patients with acute myelogenous leukemia (AML). The mutations occur either in the activation loop, for example, as point mutations of Asp835 or as internal tandem duplication (ITD) sequences in the juxtamembrane (JM) domain. To further understand the nature of FLT3 autoinhibition and regulation, we have determined the crystal structure of the autoinhibited form of FLT3. This structure shows the autoinhibitory conformation of a complete JM domain in this class of receptor tyrosine kinases. The detailed inhibitory mechanism of the JM domain is revealed, which is likely utilized by other members of type III receptor tyrosine kinases.
- Vertex Pharmaceuticals Incorporated, 130 Waverly Street, Cambridge, MA 02139, USA. james_griffith@vrtx.com
Organizational Affiliation: