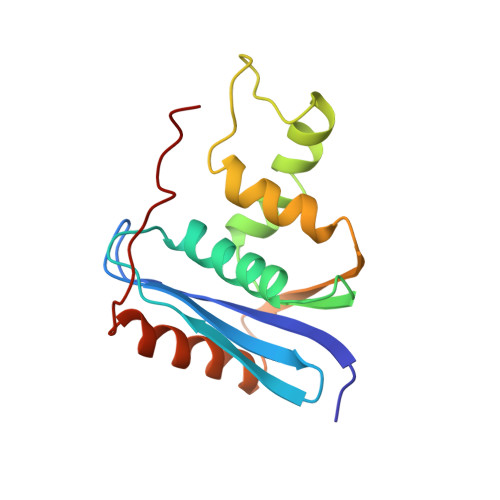
Solution Structure of Ribonuclease Hi from Escherichia Coli
Fujiwara, M., Kato, T., Yamazaki, T., Yamasaki, K., Nagayama, K.(2000) Biol Pharm Bull 23: 1147
- PubMed: 11041241
- DOI: https://doi.org/10.1248/bpb.23.1147
- Primary Citation of Related Structures:
1RCH - PubMed Abstract:
The solution structure of ribonuclease HI (RNase HI) from Escherichia coli (E. coli), a protein of 155 residues, was determined. Three-dimensional nuclear Overhauser enhancement spectroscopy (NOESY) was used to obtain 1,424 distance constraints between individually assigned polypeptide chain hydrogen atoms. Supplemental geometric constraints of 90phi angles and 12chi1 angles, and the distance constraints of 66 hydrogen bonds were experimentally derived. Using the DADAS90 program that calculates structures in dihedral angle space, 15 structures satisfying almost all constraints were obtained. The average root mean square deviation (RMSD) from the mean structure was 0.75 A for backbone atoms. The RMSD for backbone atoms between the representative NMR structure with the smallest constraint violation and crystal structures was within 1.2 A. Although the NMR and crystal structures thus resemble one another, a significant discrepancy was observed in a region termed 'basic protrusion.' The discrepancy observed in NMR experiments is explained by fluctuation in this region.
- Analytical Engineering Department. JEOL DATUM Ltd, Akishima, Tokyo, Japan. masako@jeol.co.jp
Organizational Affiliation: