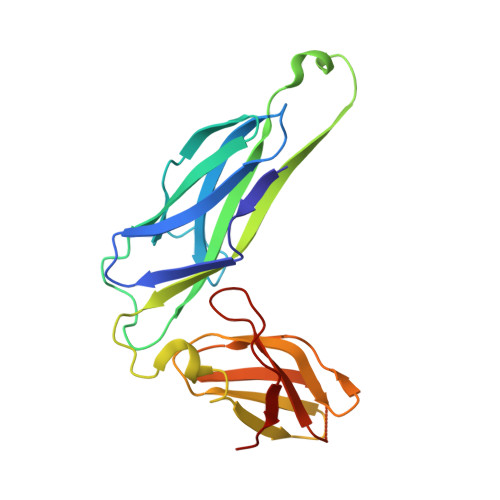
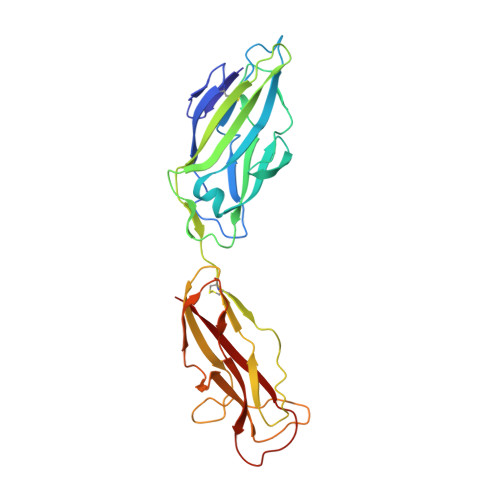
X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli.
Choudhury, D., Thompson, A., Stojanoff, V., Langermann, S., Pinkner, J., Hultgren, S.J., Knight, S.D.(1999) Science 285: 1061-1066
- PubMed: 10446051
- DOI: https://doi.org/10.1126/science.285.5430.1061
- Primary Citation of Related Structures:
1QUN - PubMed Abstract:
Type 1 pili-adhesive fibers expressed in most members of the Enterobacteriaceae family-mediate binding to mannose receptors on host cells through the FimH adhesin. Pilus biogenesis proceeds by way of the chaperone/usher pathway. The x-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli at 2.5 angstrom resolution reveals the basis for carbohydrate recognition and for pilus assembly. The carboxyl-terminal pilin domain of FimH has an immunoglobulin-like fold, except that the seventh strand is missing, leaving part of the hydrophobic core exposed. A donor strand complementation mechanism in which the chaperone donates a strand to complete the pilin domain explains the basis for both chaperone function and pilus biogenesis.
- Department of Molecular Biology, Uppsala Biomedical Center, Swedish University of Agricultural Sciences, Box 590, S-753 24 Uppsala, Sweden.
Organizational Affiliation: