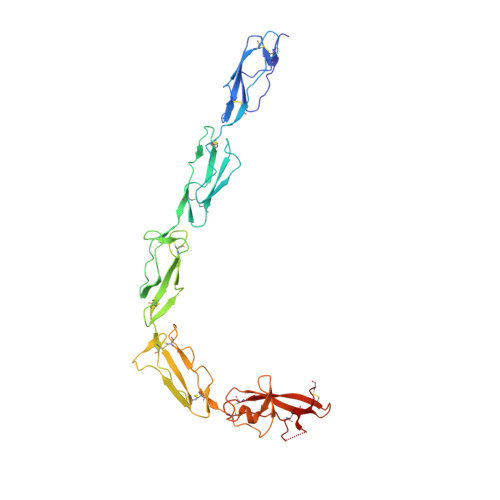
Adhesion mechanism of human beta(2)-glycoprotein I to phospholipids based on its crystal structure.
Bouma, B., de Groot, P.G., van den Elsen, J.M., Ravelli, R.B., Schouten, A., Simmelink, M.J., Derksen, R.H., Kroon, J., Gros, P.(1999) EMBO J 18: 5166-5174
- PubMed: 10508150
- DOI: https://doi.org/10.1093/emboj/18.19.5166
- Primary Citation of Related Structures:
1QUB - PubMed Abstract:
Human beta(2)-glycoprotein I is a heavily glycosylated five-domain plasma membrane-adhesion protein, which has been implicated in blood coagulation and clearance of apoptotic bodies from the circulation. It is also the key antigen in the autoimmune disease anti-phospholipid syndrome. The crystal structure of beta(2)-glycoprotein I isolated from human plasma reveals an elongated fish-hook-like arrangement of the globular short consensus repeat domains. Half of the C-terminal fifth domain deviates strongly from the standard fold, as observed in domains one to four. This aberrant half forms a specific phospholipid-binding site. A large patch of 14 positively charged residues provides electrostatic interactions with anionic phospholipid headgroups and an exposed membrane-insertion loop yields specificity for lipid layers. The observed spatial arrangement of the five domains suggests a functional partitioning of protein adhesion and membrane adhesion over the N- and C-terminal domains, respectively, separated by glycosylated bridging domains. Coordinates are in the Protein Data Bank (accession No. 1QUB).
- Department of Crystal and Structural Chemistry, Bijvoet Center for Biomolecular Research, Utrecht University, Padualaan 8, 3584 CH Utrecht, The Netherlands.
Organizational Affiliation: