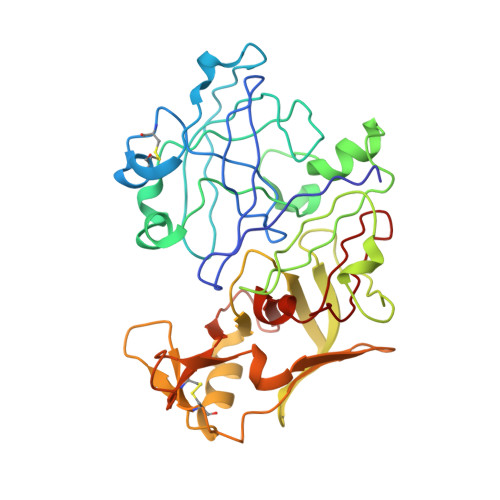
Structural insights into the activation of P. vivax plasmepsin.
Bernstein, N.K., Cherney, M.M., Yowell, C.A., Dame, J.B., James, M.N.(2003) J Mol Biology 329: 505-524
- PubMed: 12767832
- DOI: https://doi.org/10.1016/s0022-2836(03)00444-3
- Primary Citation of Related Structures:
1MIQ, 1QS8 - PubMed Abstract:
The malarial aspartic proteinases (plasmepsins) have been discovered in several species of Plasmodium, including all four of the human malarial pathogens. In P.falciparum, plasmepsins I, II, IV and HAP have been directly implicated in hemoglobin degradation during malaria infection, and are now considered targets for anti-malarial drug design. The plasmepsins are produced from inactive zymogens, proplasmepsins, having unusually long N-terminal prosegments of more than 120 amino acids. Structural and biochemical evidence suggests that the conversion process of proplasmepsins to plasmepsins differs substantially from the gastric and plant aspartic proteinases. Instead of blocking substrate access to a pre-formed active site, the prosegment enforces a conformation in which proplasmepsin cannot form a functional active site. We have determined crystal structures of plasmepsin and proplasmepsin from P.vivax. The three-dimensional structure of P.vivax plasmepsin is typical of the monomeric aspartic proteinases, and the structure of P.vivax proplasmepsin is similar to that of P.falciparum proplasmepsin II. A dramatic refolding of the mature N terminus and a large (18 degrees ) reorientation of the N-domain between P.vivax proplasmepsin and plasmepsin results in a severe distortion of the active site region of the zymogen relative to that of the mature enzyme. The present structures confirm that the mode of inactivation observed originally in P.falciparum proplasmepsin II, i.e. an incompletely formed active site, is a true structural feature and likely represents the general mode of inactivation of the related proplasmepsins.
- CIHR Group in Protein Structure and Function, Department of Biochemistry, University of Alberta, Edmonton, Alberta, Canada T6G 2H7.
Organizational Affiliation: