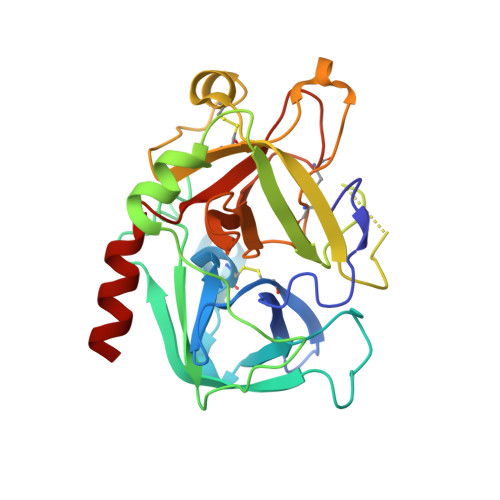
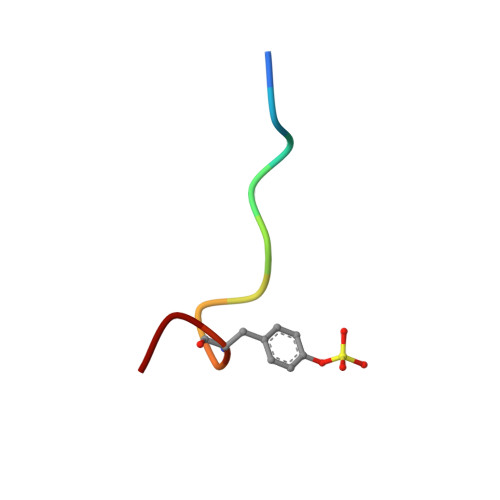
Structural analysis of thrombin complexed with potent inhibitors incorporating a phenyl group as a peptide mimetic and aminopyridines as guanidine substitutes.
Bone, R., Lu, T., Illig, C.R., Soll, R.M., Spurlino, J.C.(1998) J Med Chem 41: 2068-2075
- PubMed: 9622548
- DOI: https://doi.org/10.1021/jm970796l
- Primary Citation of Related Structures:
1QBV - PubMed Abstract:
The structure of the noncovalent complex of human alpha-thrombin with a nonpeptide inhibitor containing a central phenyl scaffold, N-[2-[5-methyl-3-(2-chlorophenylsulfonyloxy)phenoxy]ethyl]-N- methyl-4 -aminopyridine (1), has been determined to 2.20 A resolution. In addition, the thrombin-bound structures of two distinct amino acid-based inhibitors (3 and 4) containing different aminopyridine-derived guanidine mimetics have been determined. Each compound occupies the same region of the active site and projects an aminopyridine, a central hydrophobic group, and an aryl group, into the S1, S2, and aryl subsites on thrombin. Nonpeptide 1 forms only one direct intermolecular hydrogen bond to the thrombin active site and forms no hydrogen bonds to ordered molecules of solvent. Close contacts are observed between main-chain carbonyl groups on thrombin and the edges of the central phenyl and aminopyridine rings and the sulfonyl group of 1 such that atoms carrying opposite partial charges are juxtaposed. Aminopyridine groups in 3 and 4 also form close contacts with the edges of carbonyl groups on thrombin and are flexibly accommodated in the S1 subsite. Superposition of the bound conformations of 1 and D-Phe-Pro-amidobutylguanidine (2) revealed that the central phenyl scaffold of 1 substitutes for the peptide main chain of 2.
- 3-Dimensional Pharmaceuticals Inc., Eagleview Corporate Center, 665 Stockton Drive, Suite 104, Exton, Pennsylvania 19341, USA.
Organizational Affiliation: