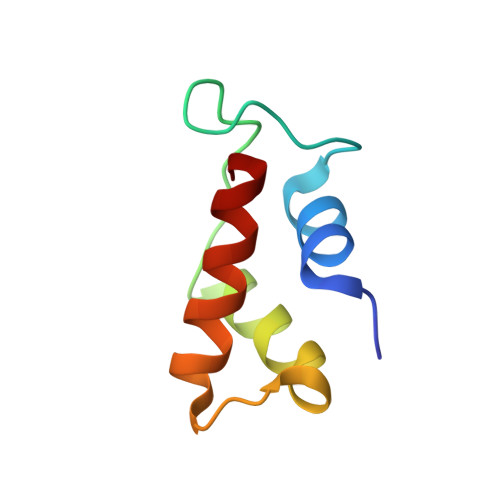
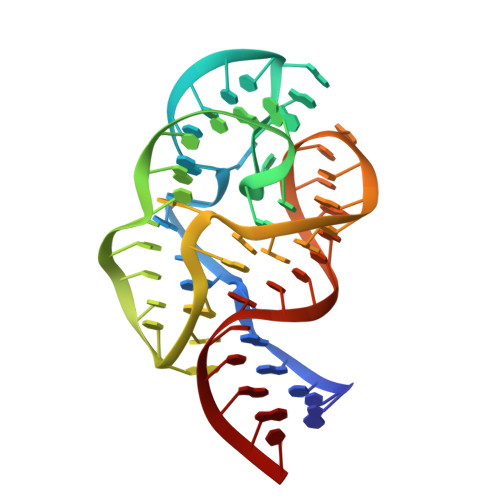
Crystal structure of a conserved ribosomal protein-RNA complex.
Conn, G.L., Draper, D.E., Lattman, E.E., Gittis, A.G.(1999) Science 284: 1171-1174
- PubMed: 10325228
- DOI: https://doi.org/10.1126/science.284.5417.1171
- Primary Citation of Related Structures:
1QA6 - PubMed Abstract:
The structure of a highly conserved complex between a 58-nucleotide domain of large subunit ribosomal RNA and the RNA-binding domain of ribosomal protein L11 has been solved at 2.8 angstrom resolution. It reveals a precisely folded RNA structure that is stabilized by extensive tertiary contacts and contains an unusually large core of stacked bases. A bulge loop base from one hairpin of the RNA is intercalated into the distorted major groove of another helix; the protein locks this tertiary interaction into place by binding to the intercalated base from the minor groove side. This direct interaction with a key ribosomal RNA tertiary interaction suggests that part of the role of L11 is to stabilize an unusual RNA fold within the ribosome.
- Department of Chemistry, Johns Hopkins University, Baltimore, MD 21218, USA.
Organizational Affiliation: