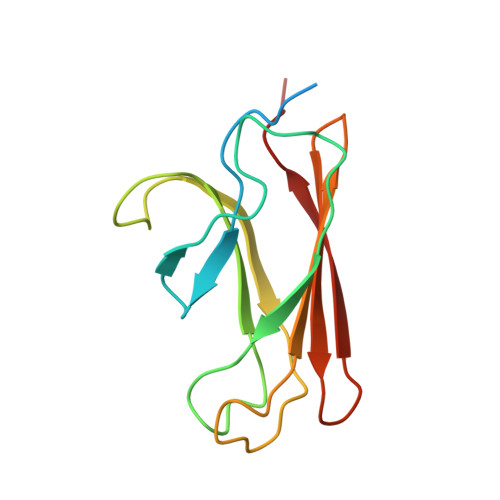
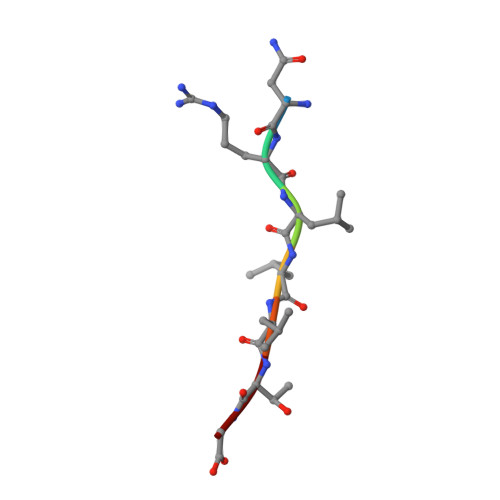
The solution structure of the bacterial HSP70 chaperone protein domain DnaK(393-507) in complex with the peptide NRLLLTG.
Stevens, S.Y., Cai, S., Pellecchia, M., Zuiderweg, E.R.(2003) Protein Sci 12: 2588-2596
- PubMed: 14573869
- DOI: https://doi.org/10.1110/ps.03269103
- Primary Citation of Related Structures:
1Q5L - PubMed Abstract:
The Hsp70 family of molecular chaperones participates in a number of cellular processes, including binding to nascent polypeptide chains and assistance in protein (re)folding and degradation. We present the solution structure of the substrate binding domain (residues 393-507) of the Escherichia coli Hsp70, DnaK, that is bound to the peptide NRLLLTG and compare it to the crystal structure of DnaK(389-607) bound to the same peptide. The construct discussed here does not contain the alpha-helical domain that characterizes earlier published peptide-bound structures of the Hsp70s. It is established that removing the alpha-helical domain in its entirety does not affect the primary interactions or structure of the DnaK(393-507) in complex with the peptide NRLLLTG. In particular, the arch that protects the substrate-binding cleft is also formed in the absence of the helical lid. 15N-relaxation measurements show that the peptide-bound form of DnaK(393-507) is relatively rigid. As compared to the peptide-free state, the peptide-bound state of the domain shows distinct, widespread, and contiguous differences in structure extending toward areas previously defined as important to the allosteric regulation of the Hsp70 chaperones.
- Biophysics Research Division, University of Michigan, Ann Arbor, Michigan 48109-1055, USA.
Organizational Affiliation: