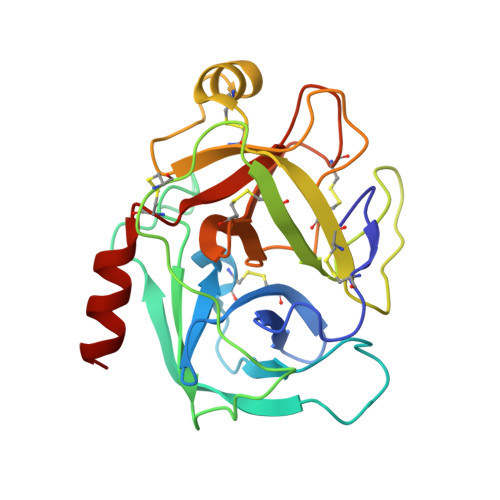
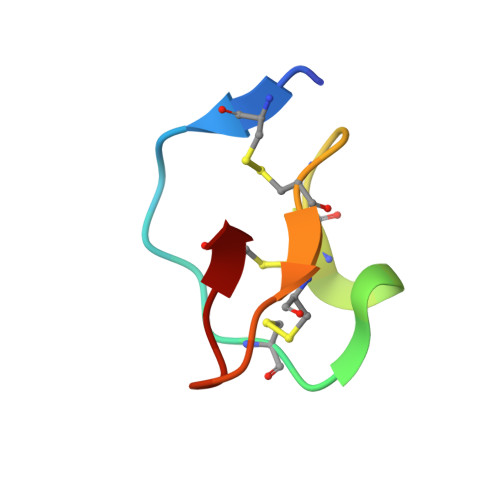
The refined 2.0 A X-ray crystal structure of the complex formed between bovine beta-trypsin and CMTI-I, a trypsin inhibitor from squash seeds (Cucurbita maxima). Topological similarity of the squash seed inhibitors with the carboxypeptidase A inhibitor from potatoes
Bode, W., Greyling, H.J., Huber, R., Otlewski, J., Wilusz, T.(1989) FEBS Lett 242: 285-292
- PubMed: 2914611
- DOI: https://doi.org/10.1016/0014-5793(89)80486-7
- Primary Citation of Related Structures:
1PPE - PubMed Abstract:
The stoichiometric complex formed between bovine beta-trypsin and the Cucurbita maxima trypsin inhibitor I (CMTI-I) was crystallized and its X-ray crystal structure determined using Patterson search techniques. Its structure has been crystallographically refined to a final R value of 0.152 (6.0-2.0 A). CMTI-I is of ellipsoidal shape; it lacks helices or beta-sheets, but consists of turns and connecting short polypeptide stretches. The disulfide pairing is CYS-3I-20I, Cys-10I-22I and Cys-16I-28I. According to the polypeptide fold and disulfide connectivity its structure resembles that of the carboxypeptidase A inhibitor from potatoes. Thirteen of the 29 inhibitor residues are in direct contact with trypsin; most of them are in the primary binding segment Val-2I (P4)-Glu-9I (P4') which contains the reactive site bond Arg-5I-Ile-6I and is in a conformation observed also for other serine proteinase inhibitors.
- Max-Planck-Institut für Biochemie, Martinsried, FRG.
Organizational Affiliation: