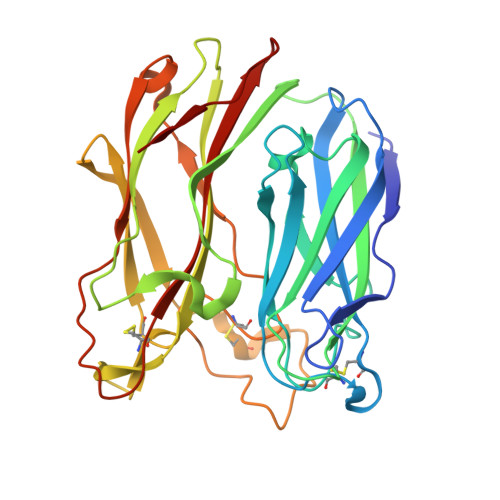
The three-dimensional structure of PNGase F, a glycosylasparaginase from Flavobacterium meningosepticum.
Norris, G.E., Stillman, T.J., Anderson, B.F., Baker, E.N.(1994) Structure 2: 1049-1059
- PubMed: 7881905
- DOI: https://doi.org/10.1016/s0969-2126(94)00108-1
- Primary Citation of Related Structures:
1PGS - PubMed Abstract:
Peptide:N-glycosidase F (PNGase F) is an enzyme that catalyzes the complete removal of N-linked oligosaccharide chains from glycoproteins. Often called an endoglycosidase, it is more correctly termed an amidase or glycosylasparaginase as cleavage is at the asparagine-sugar amide linkage. The enzyme is widely used in structure-function studies of glycoproteins. We have determined the crystal structure of PNGase F at 1.8 A resolution. The protein is folded into two domains, each with an eight-stranded antiparallel beta jelly roll configuration similar to many viral capsid proteins and also found, in expanded form, in lectins and several glucanases. Two potential active site regions have been identified, both in the interdomain region and shaped by prominent loops from one domain. Exposed aromatic residues are a feature of one site. The finding that PNGase F is based on two jelly roll domains suggests parallels with lectins and other carbohydrate-binding proteins. These proteins either bind sugars on the concave face of the beta-sandwich structure (aided by loops) or amongst the loops themselves. Further analysis of the function and identification of the catalytic site should lead to an understanding of both the specificity of PNGase F and possibly also the recognition processes that identify glycosylation sites on proteins.
- Department of Chemistry and Biochemistry, Massey University, Palmerston North, New Zealand.
Organizational Affiliation: