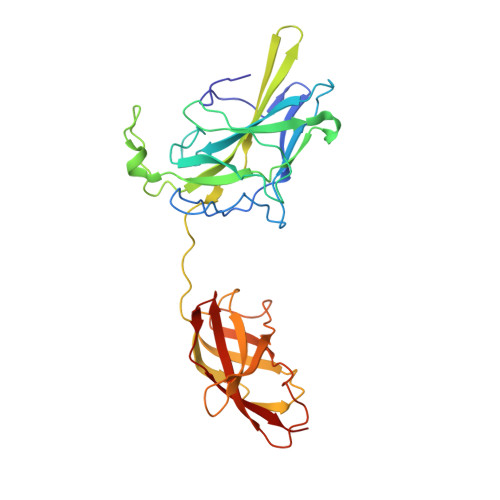
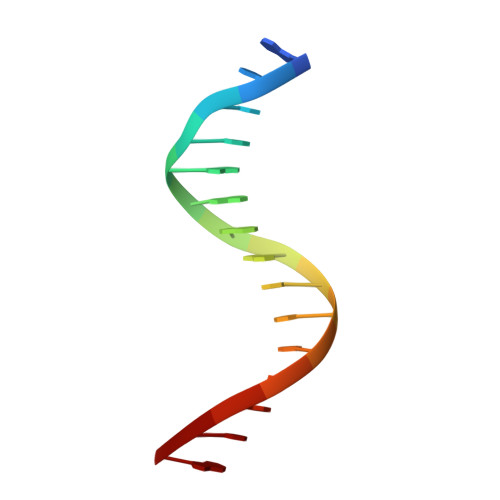
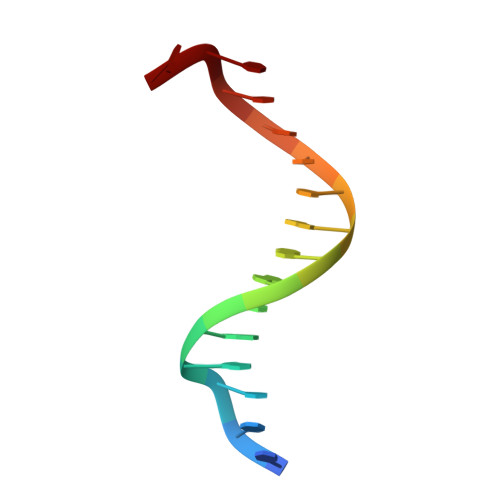
Structure of NFAT1 bound as a dimer to the HIV-1 LTR kappa B element
Giffin, M.J., Stroud, J.C., Bates, D.L., von Koenig, K.D., Hardin, J., Chen, L.(2003) Nat Struct Biol 10: 800-806
- PubMed: 12949493
- DOI: https://doi.org/10.1038/nsb981
- Primary Citation of Related Structures:
1P7H - PubMed Abstract:
DNA binding by NFAT1 as a dimer has been implicated in the activation of host and viral genes. Here we report a crystal structure of NFAT1 bound cooperatively as a dimer to the highly conserved kappa B site from the human immunodeficiency virus 1 (HIV-1) long terminal repeat (LTR). This structure reveals a new mode of dimerization and protein-DNA recognition by the Rel homology region (RHR) of NFAT1. The two NFAT1 monomers form a complete circle around the kappa B DNA through protein-protein interactions mediated by both their N- and C-terminal subdomains. The major dimer interface, formed by the C-terminal domain, is asymmetric and substantially different from the symmetric dimer interface seen in other Rel family proteins. Comparison to other NFAT structures, including NFAT5 and the NFAT1-Fos-Jun-ARRE2 complex, reveals that NFAT1 adopts different conformations and its protein surfaces mediate distinct protein-protein interactions in the context of different DNA sites.
- Department of Chemistry and Biochemistry, University of Colorado at Boulder, Boulder, Colorado 80309-0215, USA.
Organizational Affiliation: