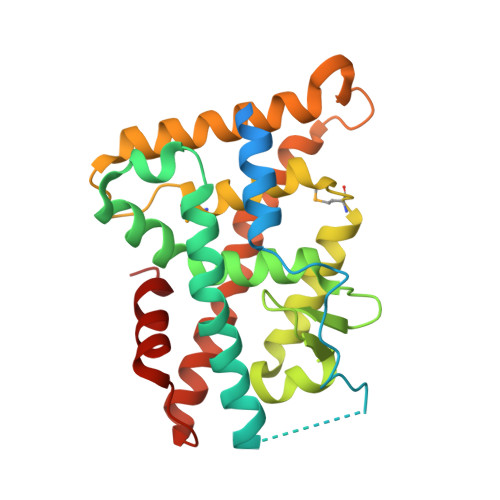
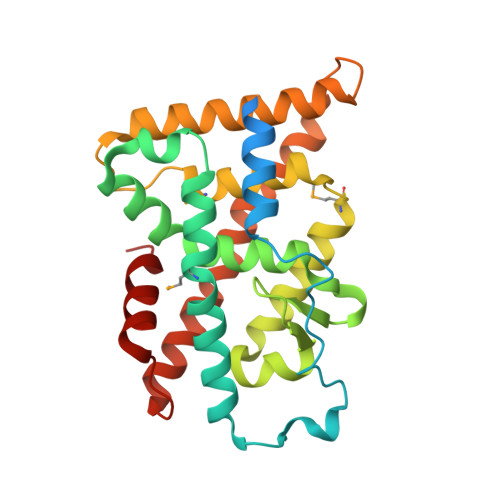
Structure and Function of Nurr1 identifies a Class of Ligand-Independent Nuclear Receptors
Wang, Z., Benoit, G., Liu, J., Prasad, S., Aarnisalo, P., Liu, X., Xu, H., Walker, N., Perlmann, T.(2003) Nature 423: 555-560
- PubMed: 12774125
- DOI: https://doi.org/10.1038/nature01645
- Primary Citation of Related Structures:
1OVL - PubMed Abstract:
Members of the nuclear receptor (NR) superfamily of transcription factors modulate gene transcription in response to small lipophilic molecules. Transcriptional activity is regulated by ligands binding to the carboxy-terminal ligand-binding domains (LBDs) of cognate NRs. A subgroup of NRs referred to as 'orphan receptors' lack identified ligands, however, raising issues about the function of their LBDs. Here we report the crystal structure of the LBD of the orphan receptor Nurr1 at 2.2 A resolution. The Nurr1 LBD adopts a canonical protein fold resembling that of agonist-bound, transcriptionally active LBDs in NRs, but the structure has two distinctive features. First, the Nurr1 LBD contains no cavity as a result of the tight packing of side chains from several bulky hydrophobic residues in the region normally occupied by ligands. Second, Nurr1 lacks a 'classical' binding site for coactivators. Despite these differences, the Nurr1 LBD can be regulated in mammalian cells. Notably, transcriptional activity is correlated with the Nurr1 LBD adopting a more stable conformation. Our findings highlight a unique structural class of NRs and define a model for ligand-independent NR function.
- Department of Structural Biology, Tularik Inc., 1120 Veterans Blvd., South San Francisco, California 94080, USA.
Organizational Affiliation: