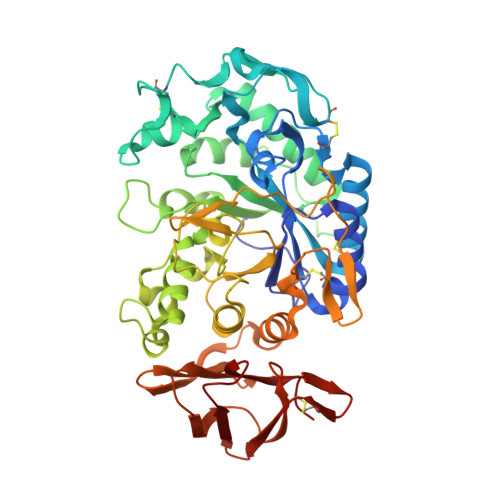
Crystal structure of pig pancreatic alpha-amylase isoenzyme II, in complex with the carbohydrate inhibitor acarbose.
Gilles, C., Astier, J.P., Marchis-Mouren, G., Cambillau, C., Payan, F.(1996) Eur J Biochem 238: 561-569
- PubMed: 8681972
- DOI: https://doi.org/10.1111/j.1432-1033.1996.0561z.x
- Primary Citation of Related Structures:
1OSE - PubMed Abstract:
Two different crystal forms of pig pancreatic alpha-amylase isoenzyme II (PPAII), free and complexed to a carbohydrate inhibitor (acarbose), have been compared together and to previously reported structures of PPAI. A crystal form obtained at 4 degrees C, containing nearly 72% solvent, made it possible to obtain a new complex with acarbose, different from a previous one obtained at 20 degrees C [Qian, M., Buisson, G., Duée, E., Haser, H. & Payan, F. (1994) Biochemistry 33, 6284-6294]. In the present form, six contiguous subsites of the enzyme active site are occupied by the carbohydrate ligand; the structural data indicate that the binding site is capable of holding more than the five glucose units of the scheme proposed through kinetic studies. A monosaccharide ring bridging two protein molecules related by the crystal packing is located on the surface, at a distance of 2.0 nm from the reducing end of the inhibitor ligand; the symmetry-related glucose ring in the crystal lattice is found 1.5 nm away from the non-reducing end of the inhibitor ligand.
- LCCMB-IBSM, CNRS, Marseille, France.
Organizational Affiliation: