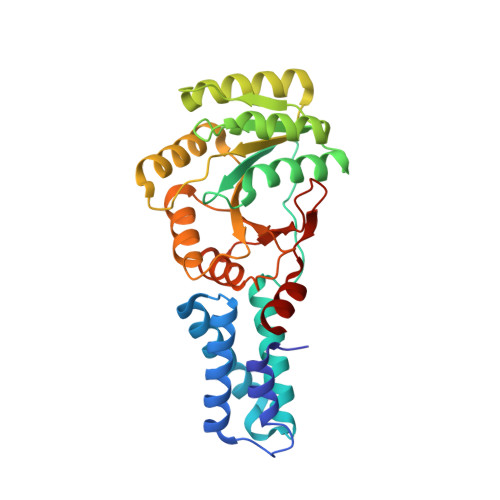
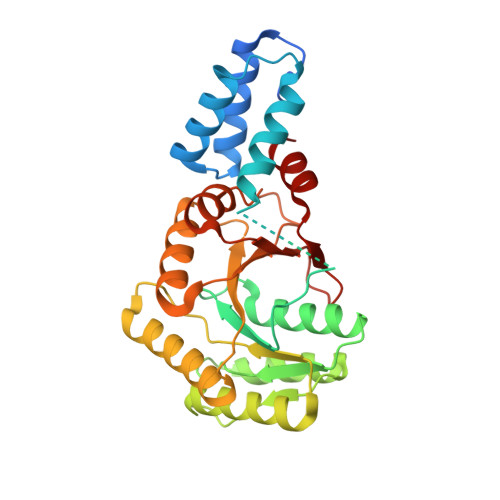
Heterodimeric Gtpase Core of the Srp Targeting Complex
Focia, P.J., Shepotinovskaya, I.V., Seidler, J.A., Freymann, D.M.(2004) Science 303: 373
- PubMed: 14726591
- DOI: https://doi.org/10.1126/science.1090827
- Primary Citation of Related Structures:
1OKK - PubMed Abstract:
Two structurally homologous guanosine triphosphatase (GTPase) domains interact directly during signal recognition particle (SRP)-mediated cotranslational targeting of proteins to the membrane. The 2.05 angstrom structure of a complex of the NG GTPase domains of Ffh and FtsY reveals a remarkably symmetric heterodimer sequestering a composite active site that contains two bound nucleotides. The structure explains the coordinate activation of the two GTPases. Conformational changes coupled to formation of their extensive interface may function allosterically to signal formation of the targeting complex to the signal-sequence binding site and the translocon. We propose that the complex represents a molecular "latch" and that its disengagement is regulated by completion of assembly of the GTPase active site.
- Department of Molecular Pharmacology and Biological Chemistry, Feinberg School of Medicine, Northwestern University, 303 East Chicago Avenue, Chicago, IL 60611, USA.
Organizational Affiliation: